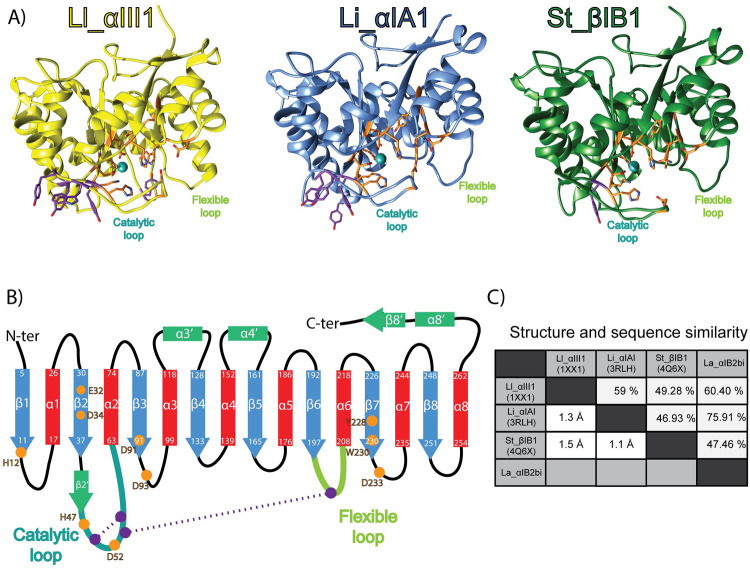
Fig 1. Overview of the SicTox PLD structures.
A) Structures of phospholipases D used in this work. Li_αIII1, Li_αIA1 and St_βIB1 are represented in yellow, blue and green, respectively. The side chains of amino acids critical for substrate binding and catalysis are shown in orange. The aromatic residues located at the putative i-face are indicated in purple. B) PLD topology: strands (blue) and helices (red) of canonical (α/β)8 barrel, other secondary structure elements (green), catalytic loop (cyan), flexible loop (green), disulphide bridges (purple). Residue numbering is based on the Li_αIA1 structure (PDB id: 3RLH). C) Structure and sequence similarity: pairwise RMSD (lower left triangle) and sequence similarity (upper right part).