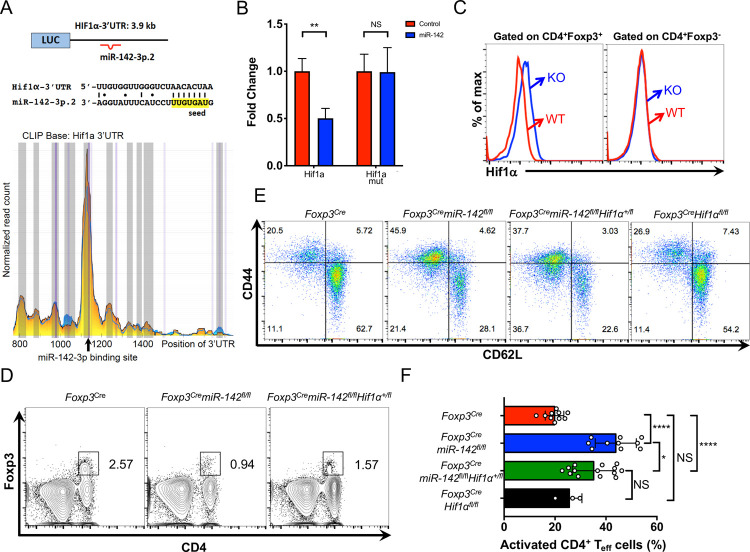
Fig 6. Lowering the Hif1a gene dose partially restores the normal size of Treg population and attenuates hyperactivation of peripheral T cells in Foxp3CremiR-142fl/fl mice.
(A) Diagram (top panel) and sequence alignment (middle panel) of miR-142-3p putative binding site in the Hif1a 3′ UTR. HITS-CLIP analysis (bottom panel) of Ago2 binding sites in the Hif1a 3′ UTR in activated CD4+ T cells (blue plot, WT; yellow plot, miR-155 KO). Note the peak of Ago2 binding activity that corresponds to the predicted miR-142-3p binding site (depicted by arrow). (B) Validation of Hif1a as direct miR-142-3p target by the 3′ UTR luciferase reporter assay. Relative expression of WT and miR-142-3p seed mutated Hif1a 3′ UTR reporter constructs upon cotransfection with either miR-142 precursor expressing plasmid or empty vector control. Expression of WT Hif1a 3′ UTR reporter in the presence of empty vector was arbitrarily set to 1. (C) Intracellular FACS analysis of Hif1α expression in CD4+Foxp3+ and CD4+Foxp3- T cells from Foxp3Cre (red line) and Foxp3CremiR-142fl/fl (blue line) spleens. (D) FACS analysis of splenic lymphocytes from 12- to 18-week-old Foxp3Cre, Foxp3CremiR-142fl/fl, and Foxp3CremiR-142fl/flHif1a+/fl mice with anti-CD4 and anti-Foxp3 specific antibodies. Foxp3+CD4+ Treg cells are gated, and numbers indicate the percentage of cells in the gate. (E) FACS analysis of CD44 and CD62L expression in splenic CD4+ T cells from Foxp3Cre, Foxp3CremiR-142fl/fl, Foxp3CremiR-142fl/flHif1a+/fl, and Foxp3CreHif1afl/fl mice. Numbers indicate percentage of cells in the quadrants. (F) Frequency of activated (CD44+CD62L−) splenic CD4+ T cells in Foxp3Cre (n = 10), Foxp3CremiR-142fl/fl (n = 10), Foxp3CremiR-142fl/flHif1a+/fl (n = 14), and Foxp3CreHif1afl/fl (n = 3) mice. Results are shown as mean ± SD. P values were calculated using 2-tailed Student t test. *, P < 0.05;**, P < 0.01; ****, P < 0.0001; NS, not significant. WT, Foxp3Cre; KO, Foxp3CremiR-142fl/fl. The underlying numerical raw data can be found in S1 Data file. The underlying flow cytometry raw data can be found at the Figshare repository. FACS, fluorescence activated cell sorting; Hif1a, hypoxia-induced factor 1 alpha; KO, knockout; SD, standard deviation; WT, wild-type.