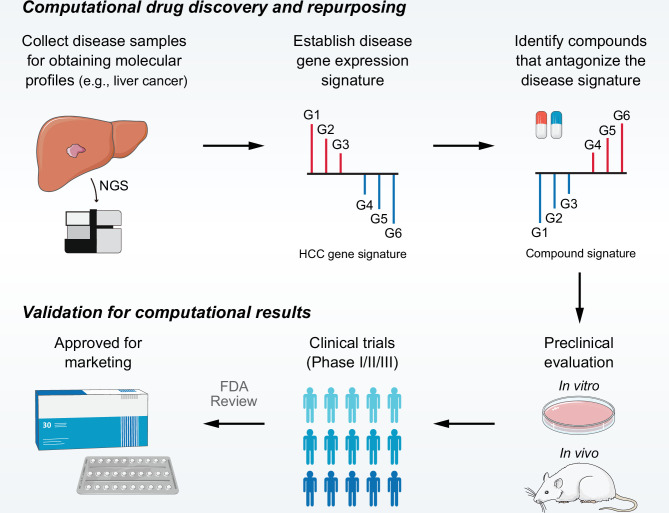
Figure 1. Overview of LINCS data-driven therapeutic discovery.
The working principle of ‘signature reversion’-based computational approach. A disease signature representing discordant expression pattern needs first to be identified (G1, G2, and G3 stand for upregulated genes while G4, G5, and G6 stand for down-regulated genes in disease state). With this signature, pharmacologic perturbation data sets can be queried to find compounds with the ability to reverse disease expression pattern (suppress expression of G1, G2, and G3 and induce expression of G4, G5, and G6). After determining the candidate compounds, experimental and clinical validation are required to translate computational findings to clinical applications. LINCS, Library of Integrated Network-based Cellular Signatures.