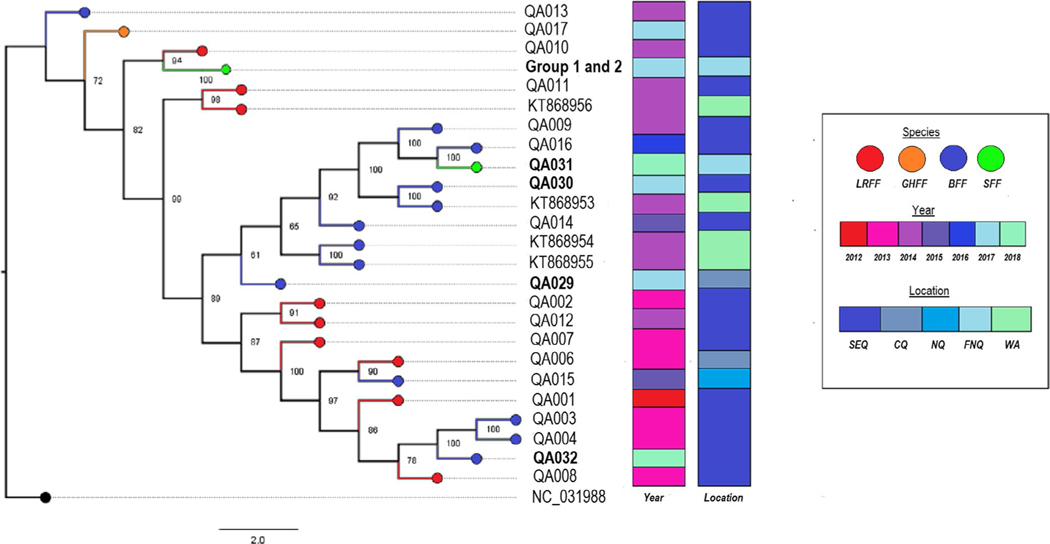
FIGURE 3.
Maximum-likelihood tree of whole genome ABLV sequences with 1,000× bootstrap support. Four ABLV whole genomes from bats in Western Australia are included for comparison, and NC_031998.1 (Gannoruwa bat lyssavirus) is used as an outgroup. The 11 sequences comprising Group 1 and Group 2 are clustered together, while the additional sequence from a P. conspicillatus pup in FNQ during the same time period as the pup cluster (QA031) is notably distant. Sequences from flying foxes submitted between November 2017 and January 2018 are in bold. aSpecies names are condensed to LRFF (little red flying fox, P. scapulatus), GHFF (grey-headed flying fox, P. poliocephalus), BFF (black flying fox, P. alecto) and SFF (spectacled flying fox, P. conspicillatus). bLocations are condensed as SEQ (South East Queensland), CQ (Central Queensland), NQ (North Queensland), FNQ (Far North Queensland) and WA (state of Western Australia)