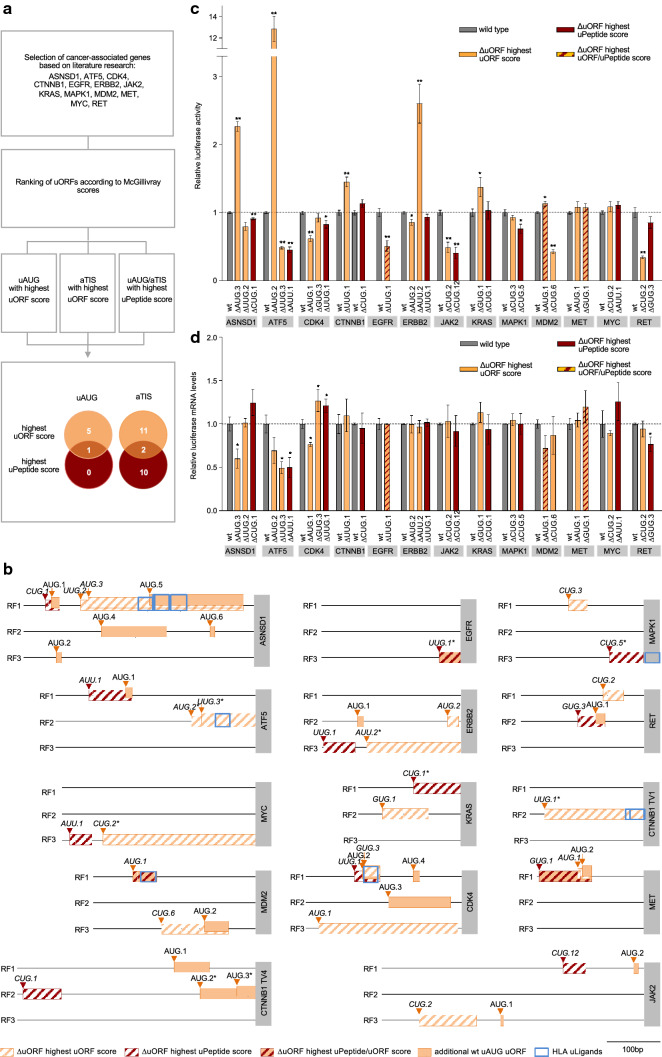
Fig. 1.
Translational regulation of CDS expression through uORFs in cancer-associated transcripts. a Flowchart illustrating the selection process for uORFs initiated at canonical (uAUG) and at alternative translational initiation sites (aTIS) for functional analysis. b Schematic illustrations of indicated in-scale TLSs displaying the analyzed uORFs (stripped boxes) and additional wild type AUG uORFs (wt, filled orange boxes) on reading frames (RF) 1–3 (black lines). Upstream ORFs overlapping into the CDS are marked with an asterisk at the respective start codon. Grey boxes on the right contain gene symbols and indicate the start of the CDS. Note that for CTNNB1 the uORFs with the highest uORF- and uPeptid-scores mapped to distinct transcript variants. Accordingly, both were selected for experimental analysis. c Bar graphs showing relative Firefly luciferase activities detected for indicated wild type (wt) and ΔuORF TLSs normalized to Renilla luciferase internal controls. Results are combined from three independent experiments and error bars indicate SEM. Levels of significance are p ≤ 0.05 (*) and p ≤ 0.01 (**) as determined by two-tailed nonparametric Mann–Whitney U-tests. d Bar graphs indicating relative Firefly luciferase mRNA levels for indicated wt and ΔuORF TLSs normalized to Renilla luciferase internal control. Results are combined from three independent experiments and error bars indicate SEM. Levels of significance are p ≤ 0.05 (*) and p ≤ 0.01 (**) as determined by two-tailed nonparametric Mann–Whitney U-tests