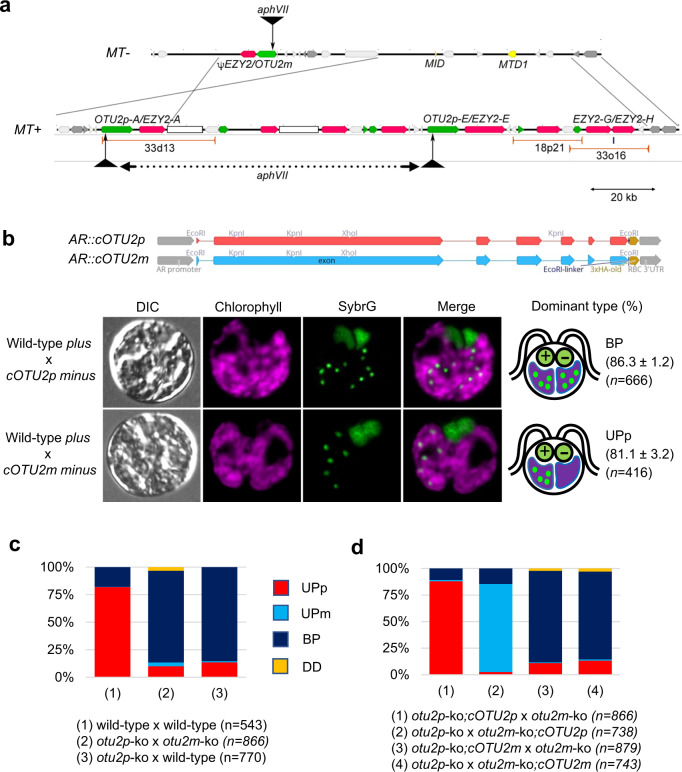
Fig. 2. MT+ -linked OTU2p establishes the uniparental inheritance of cpDNA.
a R-domain map of the mating-type loci MT− and MT+, focused on the 16-kb repeats unique to MT+. In the 16-kb repeats, OTU2 copies (green) and EZY2 copies (magenta) are alternating. BAC-plasmid clones used in this study are shown as red bars. The integration sites of aphVII delivered by CRISPR-Cas9 resulted in a ~80-kb deletion in MT+ (black triangles). b–d The average percentages of uniparental for plus (UPp), uniparental for minus (UPm), biparental (BP), and double degradation without cpDNA staining (DD) were calculated from three biological replicates. Chloroplasts from plus and minus gametes were distinguished by overlapping parent-specific mitochondrial staining of UPp vs. UPm (See Fig. 5c). “n” indicates the total number of zygotes examined. Stacked bar graphs show four cpDNA degradation patterns, whose detailed values are provided in Table S3. b Ectopic cOTU2p expression protects minus cpDNA from degradation. The above diagrams show plasmid maps of AR::cOTU2p and AR::cOTU2m. Differential interference contrast and overlaid fluorescent images of SybrG (green)-stained zygotes with chlorophyll autofluorescence (magenta) were taken at 90 min after mating. Bar = 5 μm. The percentage of the dominant cpDNA degradation pattern for each mating combination was calculated from three biological replicates (mean ± s.d.) “n” indicates the total number of zygotes examined. c Zygotes lacking OTU2p show biparental cpDNA inheritance. d Differential expression of cOTU2p between two gametes drives selective cpDNA degradation patterns in otu2-ko zygotes.