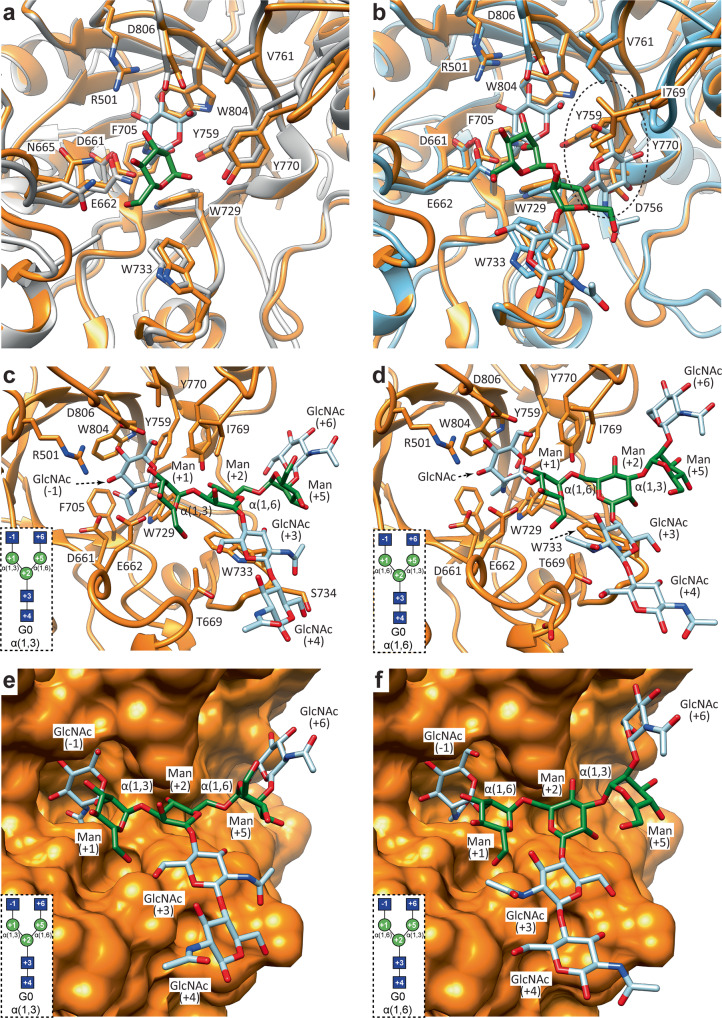
Fig. 9. Structural basis of EndoE-GH20 domain substrate specificity.
a, b Superposition of the X-ray crystal structure of EndoE-GH20 with the X-ray crystal structure of StrH-GH20A (a) (PDB code: 2YL8) and StrH-GH20B (b) (PDB code: 2YLA). Labelled residues correspond to EndoE-GH20. c, d Ribbon representation of molecular docking calculations of EndoE-GH20 with a GlcNAc2Man3GlcNAc2 substrate inserting either the α(1,3) (c) or the α(1,6) (d) antenna into the active site of the enzyme. e, f Surface representation of molecular docking calculations of EndoE-GH20 with a GlcNAc2Man3GlcNAc2 substrate inserting either the α(1,3) (e) or the α(1,6) (f) antenna into the active site of the enzyme. The schematic representations of G0 boxed in panels c–f maintain the same orientation as the cartoon representation in the corresponding panel. The carbohydrate residues are numbered based on the sugar-binding subsites in GHs88. Subsites are labeled from −n to +n (where n is an integral number); −n indicates the non-reducing end and +n the reducing end of the N-glycan. The hydrolysis takes place between −1 and +1.