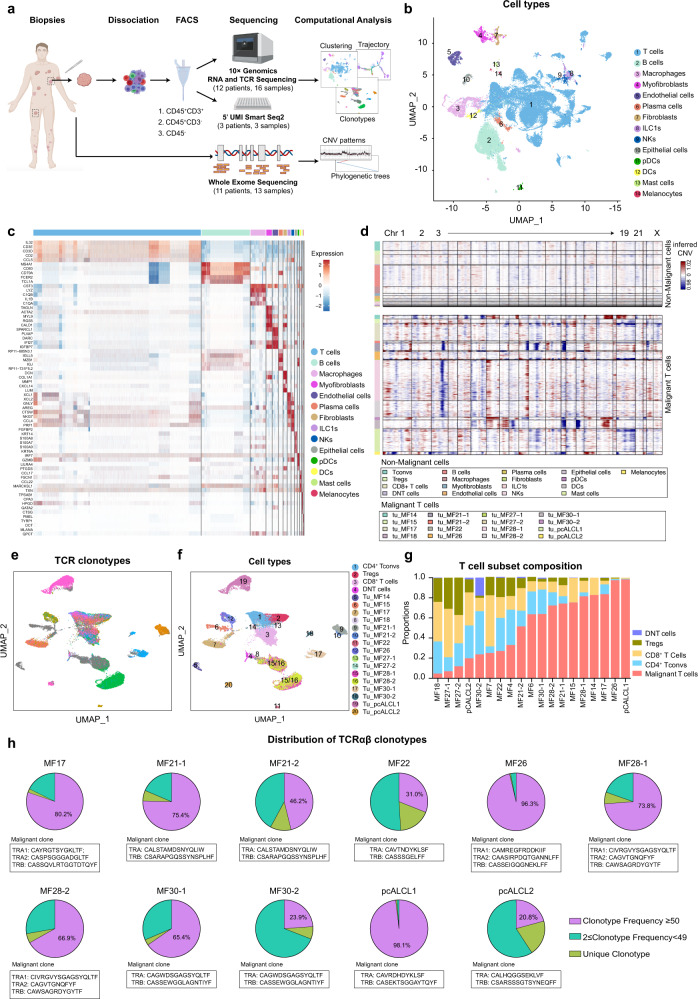
Fig. 1. Single-cell transcriptional profiling of 19 cutaneous T cell lymphoma samples.
a Workflow of tumor collection, single-cell dissociation, cell sorting, and computational analysis for scRNA-seq data and whole exome sequencing (WES) data. Among 16 samples subjected to the 10× Genomics method, the 10× 3′v3 method was applied to two samples from two patients, and the 10× 5′v2 method coupled with TCR V(D)J sequencing was applied to the remaining 14 samples from 10 patients. b UMAP plot shows 58, 926 high-quality cells from the 10× Genomics dataset. Fourteen cell types are defined by cell-specific markers. Each dot represents a single cell colored by cell type as annotated. ILC1s, type 1 innate lymphoid cells. NKs, natural killer cells. pDCs, plasmacytoid dendritic cells. DCs, dendritic cells. c Heatmap shows the expression of the top five signature genes in each cell type from the 10× Genomics dataset. Expression is indicated as the z-score normalized log2 level (count+1). d Large-scale CNVs of single cells from all samples. CNVs were inferred from the 10× Genomic dataset. e UMAP plots show all T cells from the 10× Genomics dataset after re-clustering, with cells with TCR information shown in color. Each color represents a distinct TCR clonotype. f UMAP plots show all T cells from the 10× Genomics dataset after re-clustering, with each cell colored by cell type. DNT cells: double negative T cells without expression of either CD4 or CD8B. Tu: tumor cells. Malignant T cell clusters are named by the prefix “Tu-” coupled with the sample ID. g The proportions of malignant T cells and reactive T cells in each sample. We selected 13 patients (except patients MF18 and MF27) corresponding to 16 samples with >80 malignant T cells for further malignant T cell analysis. h Pie charts show the distribution of TCRαβ clonotypes of all T cells in each sample based on clonal frequency. The paired CDR3α and CDR3β sequences with clonal frequency >50 (the dominant clonotype) in each sample are listed below the pie charts. Source data for (c) and (g) are provided in the Source Data file.