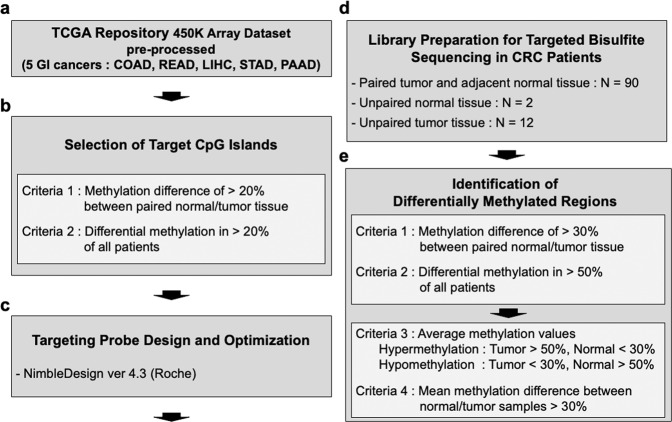
Fig. 1. Overall workflow for cohort-specific DNA methylation biomarker selection in colorectal cancer.
a Illumina Infinium 450 K array data of five major gastroenterological cancers (COAD, READ, LIHC, STAD, and PAAD) downloaded from TCGA were preprocessed. b Then, 10,754 differentially methylated CpG islands (CGIs) were shortlisted from processed 450 K array data based on our criteria. c The hybridizing probe pool targeting selected CGIs was designed using NimbleDesign. d Targeted bisulfite sequencing was conducted for 104 CRC patients from the South Korean cohort, of which 90 samples were paired tumor-adjacent healthy tissue sets, while two healthy samples and ten tumor samples were unpaired. e Generated targeted bisulfite sequencing data were analyzed to select differentially methylated regions (DMRs) in tumors relative to healthy tissues, giving rise to 40 DMRs for further examination.