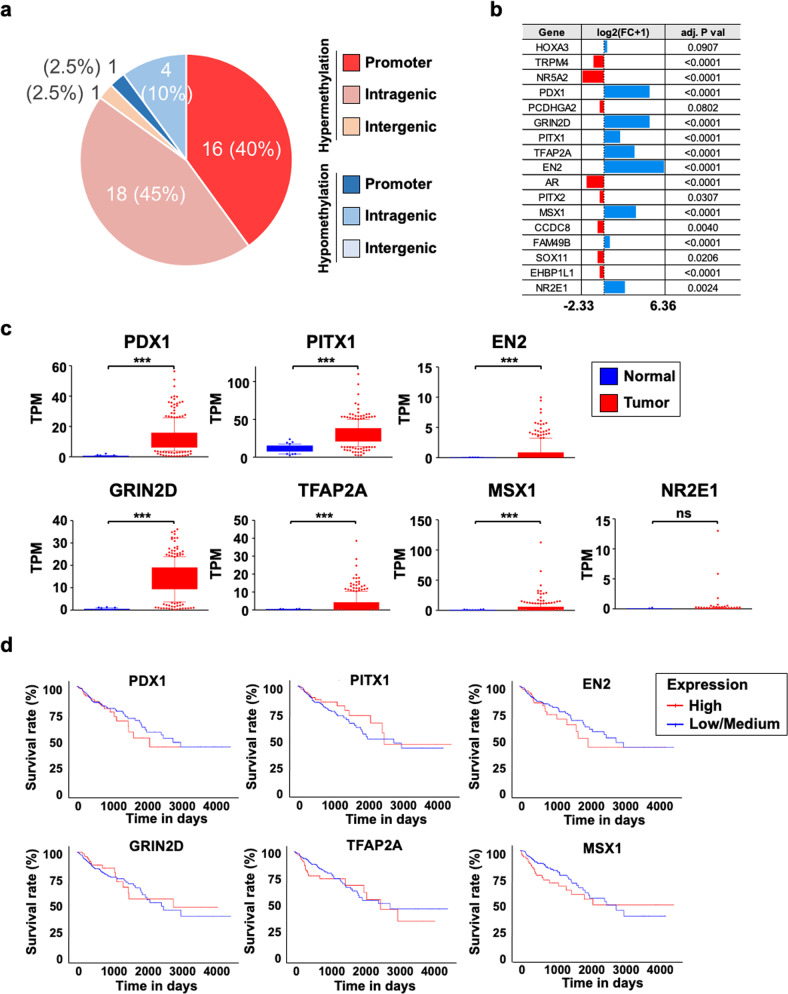
Fig. 2. Streamlining of candidate DNA methylation biomarker genes based on differential gene expression and correlation with CRC patient survival outcomes.
a Genomic location analysis of differentially methylated CGIs in targeted bisulfite sequencing data indicates that most hypermethylated regions are evenly distributed between the promoter and intragenic regions, while a larger proportion of hypomethylated regions are in intragenic regions. Our focus was on hypermethylated intragenic regions. b The expression data (read counts) downloaded from TCGA were examined to identify upregulated genes in tumor samples relative to healthy tissue samples. Downloaded RNA-seq data were processed with DESeq2 in R. c Gene expression representation of seven upregulated candidate genes in terms of TPM. Their differential expression status was further verified, and genes with nonsignificant differences were omitted from downstream analysis. Expression data between normal and tumor tissues were downloaded from TCGA, and TPM values were derived by multiplying the scaled-estimate value of RNA-seq data by 106. Significance levels are presented as ns: nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.001. d Kaplan–Meier survival plots (generated by the UALCAN database) of the six upregulated genes indicated the difference between patients with high expression of the shortlisted genes (top 25%) and patients with low or medium expression (bottom 75%). Gene expression and clinical data were based on TCGA-COAD.