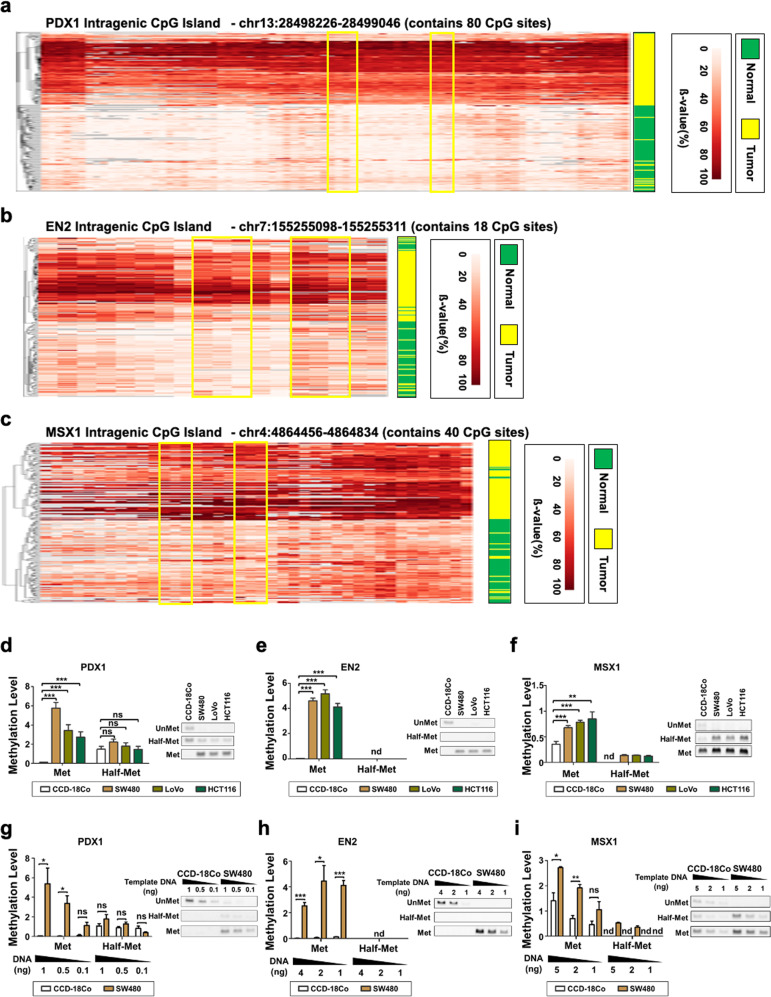
Fig. 4. Optimized benchmark for primer-binding site selection and primer design in methylation-specific PCR (MSP).
a–c MSP-targeting genomic regions in the intragenic CpG islands of PDX1 a, EN2 b, and MSX1 c are boxed in yellow. Hierarchical clustering of healthy tissue and tumor samples of targeted bisulfite sequencing data confirmed the hypermethylation of each target region in the tumor relative to healthy tissues. Each column corresponds to the cytosine of CpG sites within the respective intragenic CpG islands of PDX1, EN2, and MSX1. Low-quality sequencing data were then filtered out. d–f The efficacy of methylation detection and quantification of manually designed MSP primers were validated in vitro, in which three colon cancer cell lines (SW480, LoVo, HCT116) and one healthy colon cell line (CCD-18Co) were used. Agarose gel electrophoresis of quantitative MSP (qMSP) products also confirmed the methylation level detection efficacy of the designed primers for PDX1, EN2, and MSX1. g–i qMSP with varying CCD-18Co and SW480 template DNA quantities was conducted to verify DNA quantity-dependent signal changes of g PDX1, h EN2, and i MSX1 methylation. Met: MSP primer that binds to genomic DNA where all the target CpG sites are methylated. Half-Met: the MSP primer that binds with genomic DNA where some of the target CpG sites are methylated. Unmet: MSP primer that binds with genomic DNA where all the target CpG sites are not methylated. nd: not determined. *p < 0.05, **p < 0.01, ***p < 0.001.