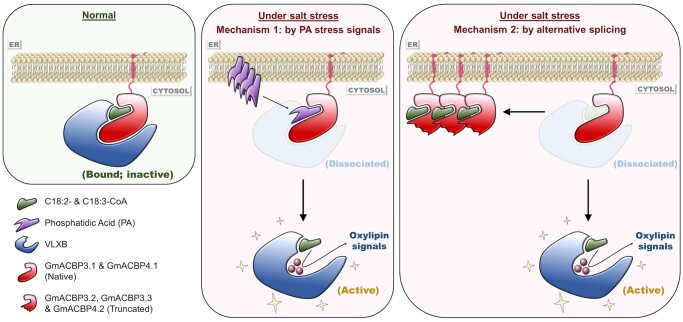
Figure 11.
Model illustrating a dual mechanism for GmACBP3/GmACBP4-mediated modulation of oxylipin signaling in salinity response. Under normal conditions (left panel), GmACBP3.1 and GmACBP4.1 (native proteins) in complex with C18:2/C18:3-CoAs interact with VLXB, which is sequestered at the ER membrane, and potentially inactivated. Under salt stress (middle and right panels), binding of GmACBP3.1 and GmACBP4.1 with these acyl-CoAs is suppressed by a dual mechanism. First, specific PA species may act as a stress signal to compete for the ACB site of GmACBP3 and GmACBP4 (middle panel). Second, alternative splicing generates variants lacking the protein–protein-interacting domain (i.e. GmACBP3.2, GmACBP3.3, and GmACBP4.2), which compete with the native proteins for acyl-CoAs (right panel). Accordingly, unbinding of C18:2/C18:3-CoAs weakens GmACBP3.1 and GmACBP4.1 interaction with VLXB, which is liberated to generate oxylipin signals and trigger salinity response. In this process, the liberated C18:2/C18:3-CoAs, upon acyl-CoA thioesterase hydrolysis to form linoleic and linolenic acids, may also serve as substrates for VLXB (middle and right panels).