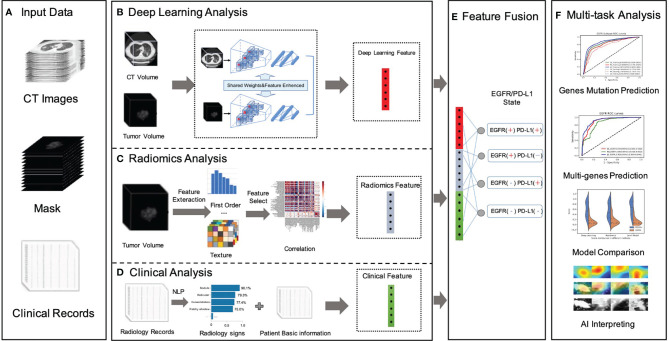
Figure 1.
Overall workflow in our study. (A) Data preparation stage included original CT image data, with manually labeled tumor images, NGS testing gene mutation status, gene mutation subtypes, radiology records, and patients’ fundamental clinical indications. (B) A novel dual-pathway deep learning network architecture that performed CT volume and tumor volume feature extraction, named DL feature, using the trained backbone for gene prediction and further to fuse the extracted feature with another learnable pathway using an asymmetric non-local fusion module. (C) The pipeline of radiomics analysis model extracted radiomics features based on manually segmented contour of the tumor. (D) The clinical feature considered a full coverage of clinical information, including radiology signs that applied NLP techniques to extract structured labels from radiology reports. (E) The feature fusion utilized full connection to provide self-adaptation on the combination of deep learning features, radiomics features, and clinical features. (F) Validation of the proposed AI system.