FIGURE 4.
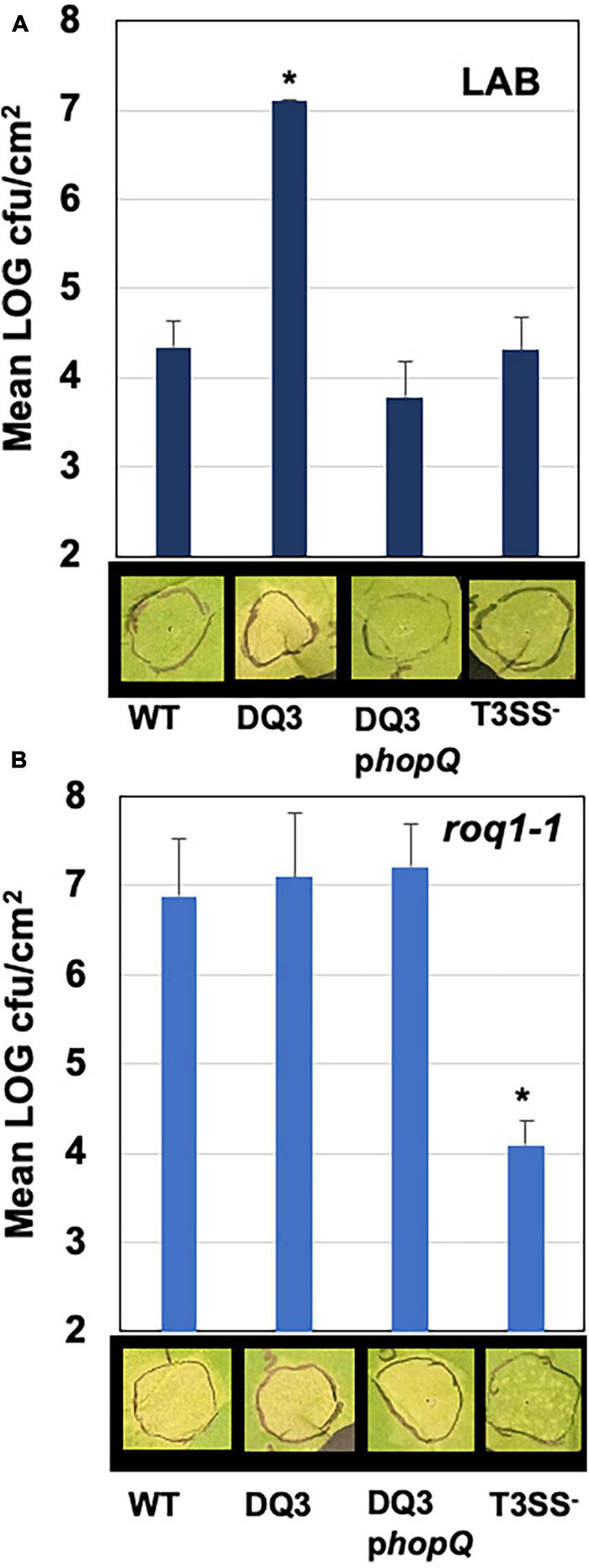
PmaES4326 gains disease compatibility with N. benthamiana in the absence of hopQ/Roq1-mediated ETI. The PmaES4326 WT strain, Pma DQ3 (ΔICE-DQ), DQ3 complemented with pCPP5372hopQ (DQ3pQ), and the non-pathogenic T3SS- strain hrcN:Tn5 were infiltrated into leaves of the N. benthamiana Lab accession (A) or the roq1-1 CRISPR mutant derivative line (B) at 3 × 104 CFU/mL. Eight days post inoculation leaves were photographed to document symptoms and bacterial load from four 4 mm diameter leaf discs from each infiltrated area by dilution plating was determined as CFUs/cm2 leaf tissue. Values are the means and standard deviations of LOG transformed CFUs/cm2 from three inoculated plants. *p < 0.05 compared to PmaES4326 WT strain as determined by unpaired one-tailed t-test.
