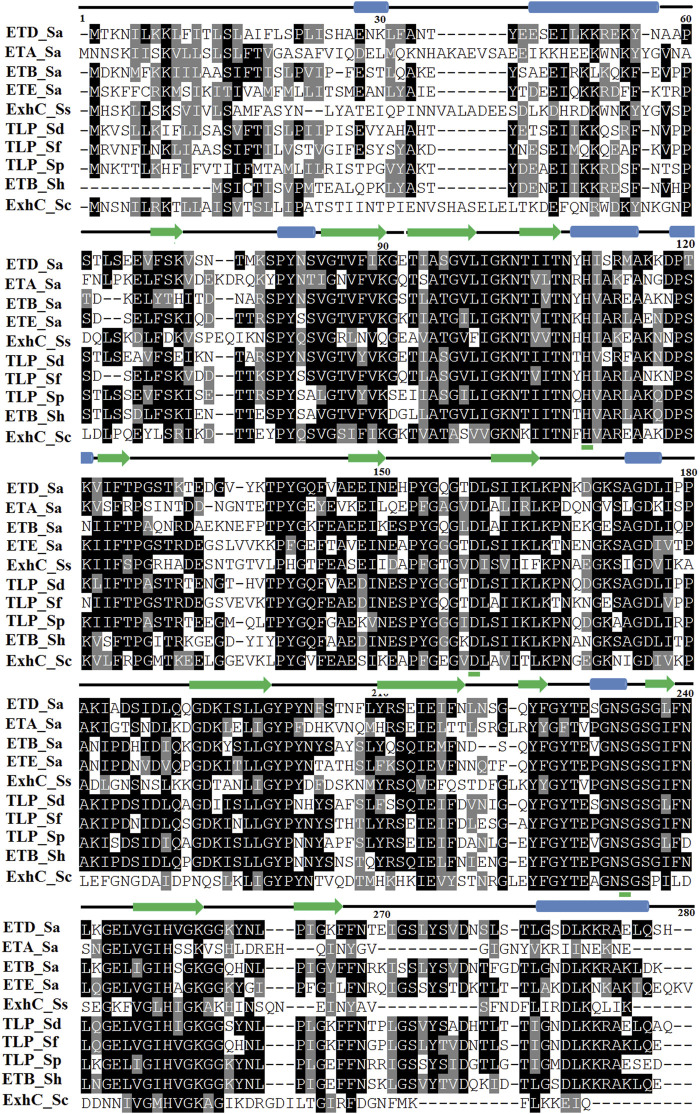
FIGURE 1.
Sequence alignment of ETs. ETD_Sa: exfoliative toxin D, Staphylococcus aureus, Gene Bank ID: AHC54578.1; ETA_Sa: epidermolytic toxin A from S. aureus, PDB ID: 1AJGJ; ETB_Sa: crystal structure of exfoliative toxin B, PDB ID: 1DT2; ETE_Sa: exfoliative toxin E from S. aureus, Gene Bank ID: WP_054190843.1, PDB ID: 5C2Z; ExhC_Ss: exfoliative toxin ExhC from S. sciuri, Gene Bank ID AEF13380.1; TLP_Sd: trypsin-like peptidase domain–containing protein from S. delphini, Gene Bank ID: WP_096546202.1; TLP_Sf: trypsin-like peptidase domain–containing protein from S. felis, Gene Bank ID: WP_103209705.1; TLP_Sp: trypsin-like peptidase domain–containing protein S. pseudintermedius, Gene Bank ID: WP_100002848.1; ETB_Sh: exfoliative toxin B from S. hyicus, Gene Bank ID: BAA99411.1, ExhB_Sc: exfoliative toxin ExhB S. chromogenes, Gene Bank ID: AAV98626.1. Amino acid residues involved in catalysis underlined with green. Secondary structural elements (alpha helices and beta strands) are shown above the sequence. Sequence numbering corresponds to the ETD_Sa precursor protein.