FIGURE 2.
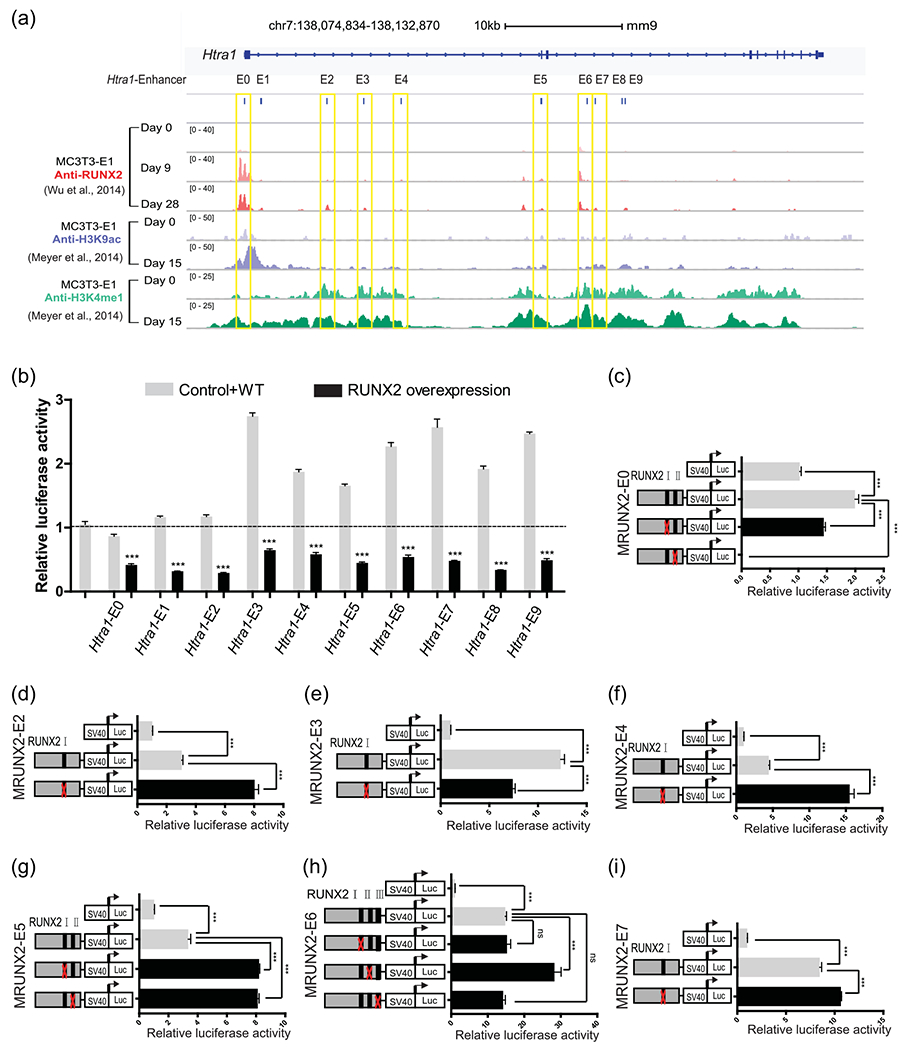
RUNX2 enrichment in 10 enhancers of the Htra1 gene locus (a) Genome browser screenshot of the Htra1 gene locus region, integrating anti-RUNX2 ChIP-Seq at osteogenic differentiation Day 0 (D0), D9, D28 in MC3T3-E1 cells (Wu et al., 2014) and anti-H3K9ac and anti-H3K4me1 at D0 and D15 in MC3T3-E1 cells (Meyer et al., 2014). MC3T3-E1 cells were cultured in osteogenic induction medium or normal growth medium. Ten putative enhancers in this locus were named Htra1-E0 to E9. The seven enhancers containing RUNX2-binding sites are marked by yellow boxes. (b) Dual luciferase assay of the 10 putative enhancers with a RUNX2 overexpression plasmid or an empty vector in HEK293FT cells. (c)–(i) Deletion analysis of RUNX2-binding sites in each putative Htra1 enhancer in MC3T3-E1 cells. All data are presented as means±SDs. *p < .05, **p < 001, ***p < .0001. Runx2, Runt-related transcription factor 2; SD, standard deviation
