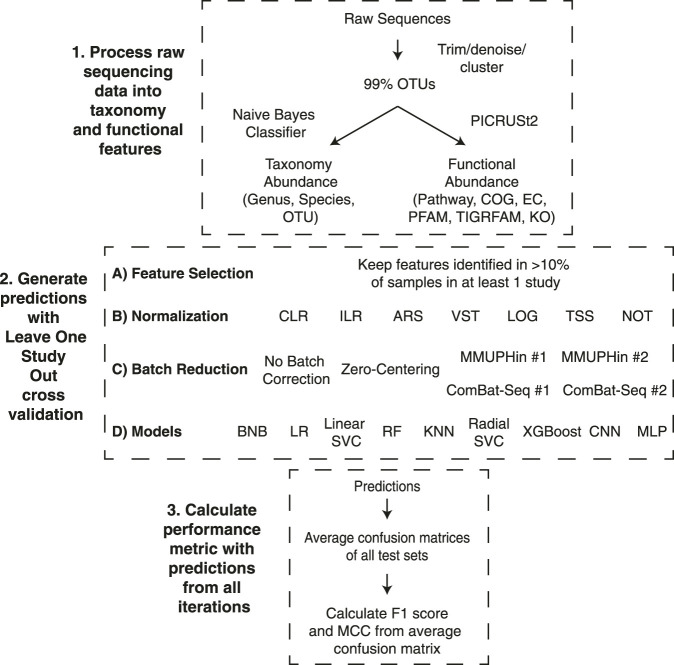
FIGURE 1.
Leave-one-dataset-out cross-validation pipeline. The experiments comprised three different stages to go from raw sequence files to the performance metrics. 1) Raw sequences were processed with Dada2 or Deblur and close-reference clustered into OTUs at 99% identity. The OTUs were classified to taxonomy at 99% confidence with a Naive Bayes classifier and used to infer functional profiles with PICRUSt2. 2) Generating predictions for the 15 iterations of our LODO cross validation consisted of all possible combinations of the listed feature selection method, normalization or transformation methods, batch effect correction methods, and models. 3) The average confusion matrix proportions across each iteration was used to generate the overall confusion matrix. The F1 Score and MCC were calculated using the proportions from the average confusion matrix. The descriptions of acronyms and abbreviations are the following: Clusters of Orthologous Groups of proteins (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (KO), Enzyme Commission (EC), Pfam protein domain (PFAM), TIGR protein family (TIGRFAM) and MetaCyc pathways (pathway), centered log-ratio (CLR), isometric log-ratio (ILR), arcsine square root transformation (ARS), variance stabilizing transformation (VST), log transformation (LOG), total sum scaling (TSS), no normalization (NOT), Bernoulli Naive Bayes (BNB), logistic regression (LR), linear support vector machine (Linear SVC), random forest (RF), K nearest neighbours (KNN), radial support vector machine (Radial SVC), eXtreme Gradient Boosting (XGBoost), convolutional neural network (CNN), multilayer perceptron (MLP).