Figure 1.
Incident SARS-CoV-2 infection associated with perturbation of blood transcriptome reflecting type 1 IFN and cell proliferation responses
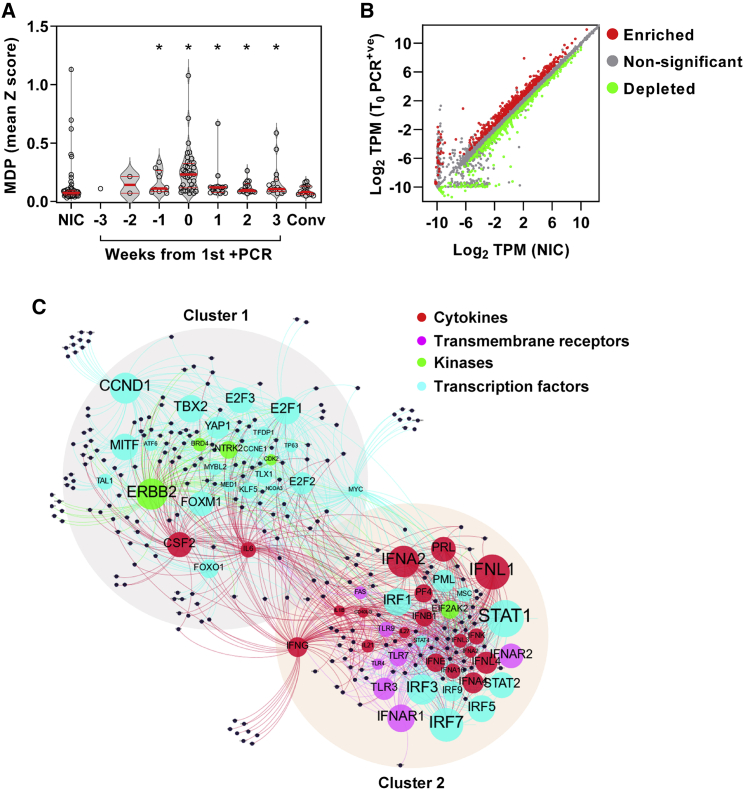
(A) Molecular degree of perturbation (MDP) in blood transcriptomes for each individual expressed as the mean of genome-wide standard deviations (Z scores) from the mean of non-infection controls (NICs). Among NICs, individuals with incident infection are stratified by weeks from first positive PCR and convalescent samples 5–6 months after incident infection. Individual data points are shown with violin plots depicting median, IQR, and frequency distributions (∗FDR < 0.05 by Kruskal-Wallis test for each group compared with NIC).
(B) Differentially expressed genes in blood transcriptomes at time of first positive PCR (T0_PCR+ve) compared with NICs. (TPM, transcripts per million).
(C) Predicted upstream regulators (labeled nodes) stratified by molecular function for differentially expressed genes (black nodes). Size of the nodes for upstream regulators is proportional to -Log10 p value. Nodes were clustered using Force Atlas 2 algorithm in GEPHI (version 0.9.2).