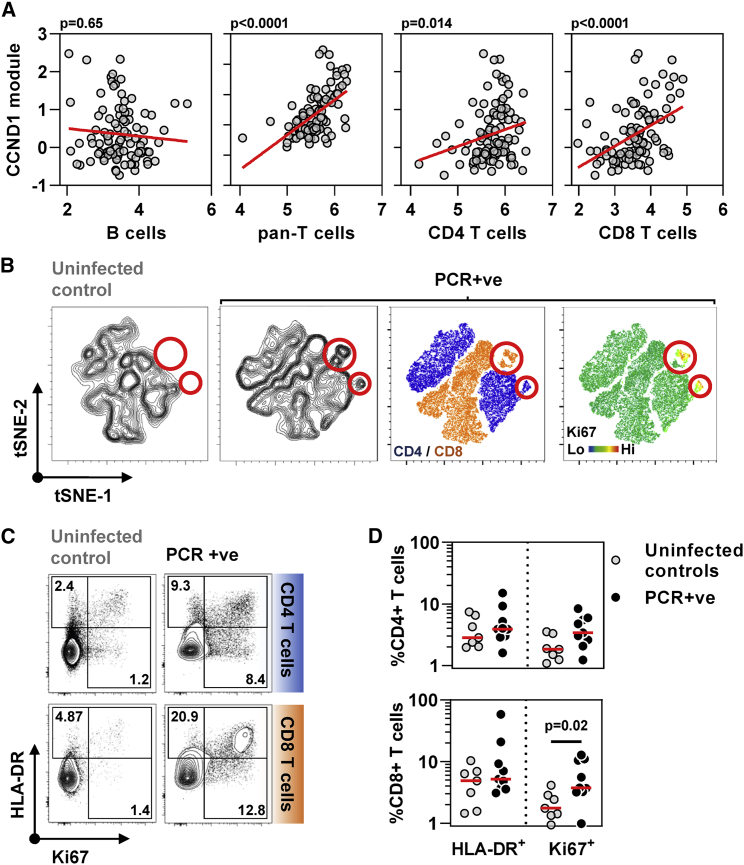
Figure 3.
Cell proliferation response to SARS-CoV-2 infection in blood transcriptomic data is attributable to T cell proliferation
(A) Correlation of CCND1 module (representative of cell proliferation response) in all time points (−3 to +3 weeks) from individuals with SARS-CoV-2 infection with each of blood transcriptomic modules representative of B cells, pan-T cells, CD4 T cells, and CD8 T cells (regression lines shown in red, p values for Spearman rank correlations).
(B) tSNE plots of T cells from non-infected controls or individuals with co-incident PCR-positive SARS-CoV-2 infection. Contour plots are shown in the two left-hand panels, followed by dot plots colored by CD4/CD8 staining or relative Ki67 staining as a proliferation marker. Red circles highlight a population of Ki67 high CD4 and CD8 cells exclusive to the PCR+ group (tSNE derived from flow cytometry data of CD4 and CD8 T cells, seven NICs, and nine PCR-positive).
(C) Representative flow cytometry data for HLA-DR and Ki67 staining in either CD4 T cells or CD8 T cells from one non-infected control and one SARS-CoV-2 infected individual at the time of the first positive PCR infection. Numbers indicate percent positive for each marker including double-positives.
(D) Summary HLA-DR and Ki67 staining data and median from seven uninfected controls and nine individuals with co-incident infection in either CD4 T cells or CD8 T cells. p value shown for Mann-Whitney test.