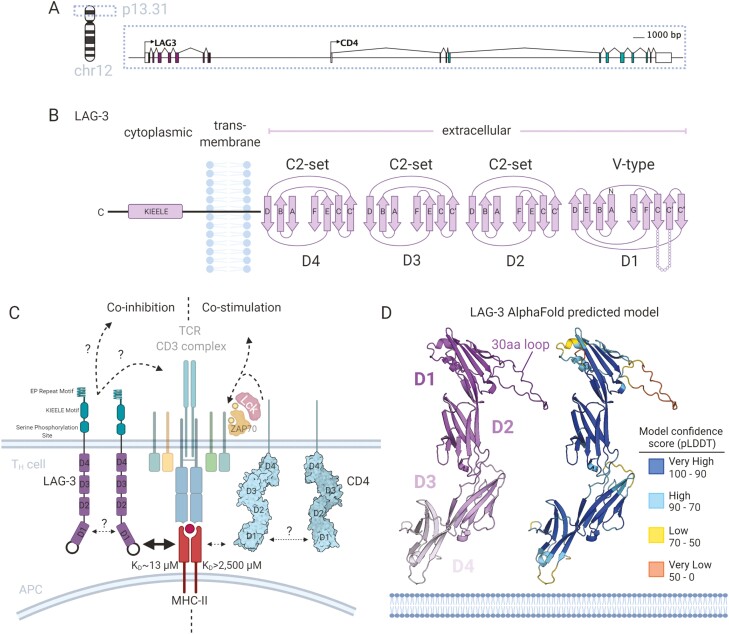
Figure 2.
Molecular understanding of LAG-3 binding to MHC-II as a co-inhibitory competitor to CD4. (A) Gene structure and location of LAG-3 on chromosome 12 adjacent to CD4. (B) The predicted protein domain folds of LAG-3 containing four extracellular Ig-domains (D1–D4) of V-type (D1) and C2-set (D2–D4). LAG-3 has a single-pass transmembrane domain and a short cytoplasmic tail containing a unique KIEELE motif involved in signal transduction. (C) Model of LAG-3 co-inhibitory competition with CD4 whereby LAG-3 binds MHC-II at a higher affinity than the co-stimulatory CD4 and transducing inhibitory signalling through undescribed signalling pathways. (D) Model of the LAG-3 extracellular domain as predicted by the AlphaFold project. The LAG-3 model is shown as cartoon representation and coloured (left) by domain (colours indicated inset) and (right) model confidence score (pLDDT) with colours indicated by the inset legend table. pLDDT confidence levels and definitions are as described previously [31].