Figure 1.
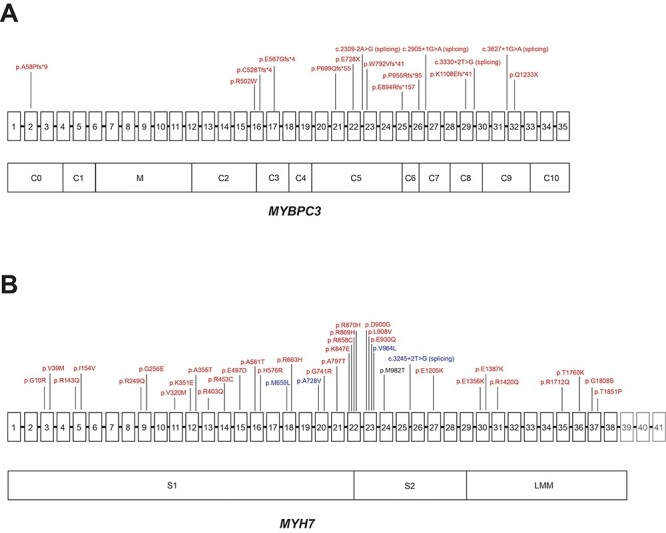
Distribution of disease-associated variants in MYH7 and MYBPC3. (A) Schematic of MYBPC3 gene with exons 1–35 (exons not to scale) above and domains below, with variants labeled in red denoting amino acid change (or location of splice variant) for pLOF or adjudicated missense variants that were associated with HCM in PMBB. (B) Schematic of MYH7 gene with exons 1–41 (exons not to scale) above and domains below. Variants are labeled in red denoting amino acid change for pDM variants with REVEL ≥ 0.5, which were associated with HCM in PMBB, and variants are labeled in blue denoting amino acid change (or location of splice variant) for pLOF or pDM variants with REVEL ≥ 0.5, which were associated with ‘Muscular wasting and disuse atrophy’ in PMBB. Note: exons 39–41, which do not encode for protein, are grayed out. Note 2: p.M982T in MYH7 was associated with both HCM and muscular wasting and disuse atrophy, but via different individuals, and is thus labeled in black.
