Abstract
Observational evidence links higher blood levels of copper with higher risk of cardiovascular diseases. However, whether those associations reflect causal links or can be attributed to confounding is still not fully clear. We investigated causal effects of copper on the risk of cardiometabolic endpoints (stroke, coronary artery disease [CAD] and type 2 diabetes) and cardiometabolic risk factors in two-sample Mendelian randomization (MR) studies. The selection of genetic instruments for blood copper levels relied on meta-analysis of genome-wide association studies in three independent studies (European Prospective Investigation into Cancer and Nutrition-Potsdam study, Prospective investigation of the Vasculature in Uppsala Seniors study, Queensland Institute of Medical Research studies). For the selected instruments, outcome associations were drawn from large public genetic consortia on the respective disease endpoints (MEGASTROKE, Cardiogram, DIAGRAM) and cardiometabolic risk factors. MR results indicate an inverse association for genetically higher copper levels with risk of CAD (odds ratio [95% confidence interval] = 0.92 [0.86–0.99], P = 0.022) and systolic blood pressure (beta [standard error (SE)] = −0.238 [0.121]; P = 0.049). Multivariable MR incorporating copper and systolic blood pressure into one model suggested systolic blood pressure as mediating factor between copper and CAD risk. In contrast to previous observational evidence establishing higher blood copper levels as risk-increasing factor for cardiometabolic diseases, this study suggests that higher levels of genetically predicted copper might play a protective role for the development of CAD and systolic blood pressure.
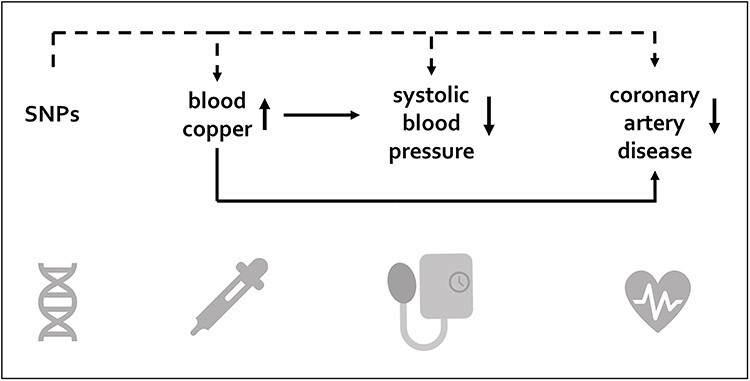
Graphical Abstract
Graphical Abstract.
Introduction
Blood levels of copper have been associated with higher risk of chronic diseases such as cardiovascular disease (CVD) and stroke in several observational studies (1,2). Although serum concentrations of copper are strictly regulated by compensatory mechanisms within certain ranges of nutritional intake, under inflammatory conditions, aging or age-related disorders inducing oxidative stress, serum concentrations of copper are increased (3,4). However, it is still not fully clear whether these changes might causally impact disease risk.
To investigate causal associations between risk factors and disease outcomes, genetic variants can be used as instrumental variables in Mendelian randomization (MR). The principle is based on Mendel’s second law of inheritance, stating that the assignment of alleles is random during meiosis. Therefore, genetic variants are generally not associated with possible confounders (e.g. environmental exposures or lifestyle factors) in exposure-disease associations (5). A previous MR study found an inverse association between higher blood copper levels and risk of ischemic heart disease (IHD) (odds ratio [OR]: 0.94; 95% confidence interval [CI]: 0.90, 0.98) (6). However, there were no MR studies on other cardiometabolic disease outcomes. Therefore, we first aimed to use human genetics to study the associations of blood copper on the risk of type 2 diabetes, coronary artery disease (CAD) and stroke. Second, we investigated related risk factors that may mediate associations between copper and disease endpoints.
Results
Selection of genetic instruments by meta-analysis of GWAS in EPIC-Potsdam, PIVUS and QIMR
We conducted genome-wide association study (GWAS) on serum copper levels within European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam data (Supplementary Material, Fig. S1) and meta-analyzed these data with published GWAS datasets on blood copper levels (7,8) (Supplementary Material, Fig. S2). We identified two genome-wide significant hits for blood copper levels. One was located in an intergenic region near selenium binding protein 1 (SELENBP1; lead single nucleotide polymorphism [SNP]: rs17564336). The other was located in the intronic region of CP (rs34951015) (Table 1, Supplementary Material, Table S1). Both instruments had F-statistics higher than 10. A complete list of genome-wide significant SNPs can be found in Supplementary Material, Table S2.
Table 1.
Independent GWAS hits for copper at genome-wide significance (< 5 × 10−08) from Meta-analysis of EPIC-Potsdam, PIVUS and QIMR
| SNP | Chr:bp | Consequence/Gene | CADDa | Effect allele | Other allele | N | EAF | P-value | Directionb | F-statistic (58) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17564336 | 1:151349174 | Intergenic/near SELENBP1 | 1.751 | T | G | 6937 | 0.298 | 1.79 × 10−17 | −−− | 73.54 |
| rs34951015 | 3:148926315 | Intronic/CP | 0.419 | A | G | 6937 | 0.099 | 9.57 × 10−10 | +++ | 37.81 |
CADD, combined annotation dependent depletion; EAF, effect allele frequency.
Extracted from Ensembl database (11).
From three different studies: EPIC-Potsdam, PIVUS and QIMR.
Causal estimates for copper on risk of chronic diseases and cardiometabolic risk factors
We could not find indication for a causal effect of copper levels on risk of type 2 diabetes or stroke (Tables 2, Supplementary Material, Table S3 and S4, Supplementary Material, Fig. S3). However, we found a significant inverse association for copper and CAD (beta [SE] = −0.079 [0.035], OR [95% CI] =0.92 [0.86–0.99], P = 0.022; Tables 2, Supplementary Material, Table S4, Supplementary Material, Fig. S3).
Table 2.
Causal estimates for copper and cardiometabolic disease endpoints and related risk factors from MR analysis
| Outcome | IVWa | Cochran’s Q statistic (P-value) |
|---|---|---|
| Type 2 diabetes | 0.028 (0.026), 0.275 | 2.814 (0.093) |
| Coronary artery disease | −0.079 (0.035), 0.022 | 2.215 (0.137) |
| Stroke | 0.034 (0.038), 0.366 | 2.233 (0.135) |
| BMI | 0.009 (0.008), 0.238 | 11.794 (0.001) |
| LDL | 0.003 (0.009), 0.729 | 2.221 (0.136) |
| HDL | −0.001 (0.009), 0.928 | 2.537 (0.111) |
| Systolic blood pressure | −0.238 (0.121), 0.049 | 0.538 (0.463) |
| Diastolic blood pressure | −0.075 (0.069), 0.280 | 0.044 (0.833) |
| HbA1cb | −0.007 (0.021), 0.728 | -- |
| Blood glucose | 0.004 (0.010), 0.661 | 0.006 (0.936) |
Values are beta (SE); P-values are from fixed effects models.
Wald ratios as only one SNP (rs17564336) was available in the outcome dataset.
In MR analyses of copper and cardiometabolic risk factors, we found indication for an inverse association with systolic blood pressure (beta [SE] = −0.238 [0.121]; P = 0.049; Tables 2, Supplementary Material, Table S3 and S4, Supplementary Material, Fig. S3).
Multivariable MR indicated a direct causal effect of systolic blood pressure on CAD (beta [SE] = 0.034 [0.003], OR [95% CI] =1.03 [1.03–1.04], P = 1.81 × 10−36), whereas for copper no significant association was observed after adjusting for systolic blood pressure (beta [SE] = −0.019 [0.027], OR [95% CI] =0.98 [0.93–1.04], P = 0.4935). (Fig. 1, Supplementary Material, Table S5) There was indication for weak instruments for copper with conditional F-statistic of 1.18. When we corrected for weak instruments in the multivariable MR, we received comparable estimates for systolic blood pressure (beta [95% CI] = 0.034 [0.027–0.041], OR [95% CI] =1.04 [1.03–1.04] and copper (beta [95% CI] = −0.028 [−0.091–0.025], OR [95% CI] =0.97 [0.91–1.03]).
Figure 1.
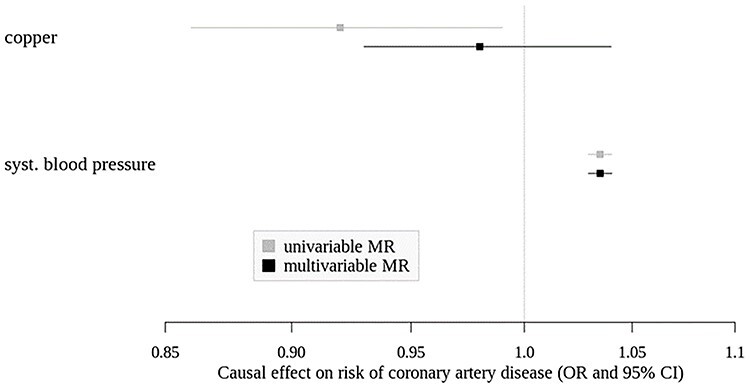
Causal effects (OR and 95% CI) of copper and systolic blood pressure on risk of coronary artery disease from univariable and multivariable MR. Estimate for copper from univariable MR from fixed effect IVW model.
Discussion
Our two-sample MR approach indicated an inverse association of genetically higher copper on risk of CAD. Similarly, we obtained an inverse association for genetically higher copper on systolic blood pressure. Multivariable MR suggested systolic blood pressure as mediating factor between copper and CAD risk.
Copper and cardiometabolic risk factors and diseases
Prospective studies on copper levels and cardiometabolic outcomes are scarce. In EPIC-Potsdam, we found a positive association between serum copper and risk of CVD and type 2 diabetes in an observational setting, the latter, however, not significant (1). Other studies showed positive associations between copper levels and all-cause mortality (9) (10) or CVD mortality (10). A prospective study in Chinese Adults indicated a positive association with risk of stroke (2). In contrast to observational studies, a previous MR study linked higher copper levels to lower risk of IHD (6). In the present MR, we could replicate this finding using a slightly different genetic instrument. Indeed, Kodali et al. used also an intergenic instrument near SELENBP1 (rs2769270, LD to our variant rs17564336: r2 = 0.43, D′ = 0.99 (11)) for copper, that—similarly to our results—drove the inverse association with IHD in their study (6). The other instrument (rs1175550) that they used was also identified by Evans et al. (7) in the Queensland Institute of Medical Research (QIMR) dataset. However, this variant was not replicated in Prospective investigation of the vasculature in Uppsala seniors (PIVUS) and EPIC-Potsdam and therefore, we did not use it for the current MR study. Although there are shared etiological pathways between CAD and other cardiometabolic diseases, especially stroke; we found no significant association of higher copper levels with risk of stroke or type 2 diabetes.
One explanation might be due to survival selection bias due to competing risks, where people die earlier from CAD than from stroke or type 2 diabetes and are hence not recruited for studies on the latter endpoints (12).
Besides cardiometabolic diseases, we also investigated related risk factors that might act as mediators between copper levels and disease risk. In this regard, a previous MR found inverse associations between copper levels and different disorders of lipid metabolism (hyperlipidemia, hypercholesterolemia) and lipid biomarkers (low-density lipoprotein (LDL), total cholesterol) using instruments from Evans et al. (7) and outcome data from the Global Lipids Genetics Consortium (13). Similar to our results, the latter associations of copper and lipid biomarkers could not be observed when using data from the UK Biobank (13). Another important risk factor for CAD is elevated blood pressure/hypertension. Observational studies found diverse associations between blood copper values and blood pressure, supporting both directions (14). Our univariable MR analyses suggest inverse associations of higher copper levels on systolic blood pressure and diastolic blood pressure, the latter, although, not significant. In multivariable MR, we found a direct effect for systolic blood pressure on CAD risk, attenuating the association between copper and CAD risk.
Biological mechanisms linking copper to CAD and systolic blood pressure
In the circulation, 80–95% of total blood copper is bound to ceruloplasmin (Cp) with high affinity. Therefore, increased blood levels of copper may be a consequence of increased transcription of Cp in liver tissues (3). One of our genetic instruments (rs34951015) is located in the Cp encoding CP gene region, suggesting that besides copper itself, also its transport protein might be involved in biological pathways linking copper to cardiometabolic risk. The SNP represents a novel hit for blood copper levels and is in LD to a potentially functional CP variant in the promoter region upstream of the CP gene, previously associated with Cp levels (rs11708215: r2 = 0.35, D′ = 0.99 (11)) (15). Cp was associated with increased risk for myocardial infarction (16), CVD mortality (10,17) and stroke (17) in observational setting and higher risk of arterial fibrillation (15) in MR approach. Based on GTEx data, CP is mainly expressed in liver, artery-aorta and artery-tibial tissues (18). Our CP instrument rs34951015 represents an eQTL for CP in artery-tibia (P = 1.3 × 10−4) and artery-aorta (P = 0.007) tissues (18).
The other instrument (rs17564336) is located near the SELENBP1 gene and represents an eQTL for SELENBP1 in artery-tibia (P = 9.2 × 10−21) and artery- aorta (P = 1.4 × 10−4) tissues (18). Besides selenium, SELENBP1 may also bind copper (19). Extracellular SELENBP1 was identified in blood after myocardial infarction or during cardiac surgery (20,21).
Our instruments show furthermore associations with red blood cell traits (rs17564336) (22) and histidine (rs34951015) (22), that may convey vertical pleiotropic pathways on the way to CAD development (23,24). Indeed, we found inverse associations between copper and systolic blood pressure which might represent a mediating pathway between copper levels and the reduction of CAD risk. Copper is an essential trace element acting as co-factor for several copper-dependent enzymes e.g. lysyl oxidase and copper-zinc superoxide dismutase that convey important biological functions like cross-linking of collagen and elastin in blood vessels and protection against free radicals, respectively. Hence, copper-deficiency has been reviewed in the context of development of CVD (25,26), dysregulation of microcirculation (27) and impaired lipid metabolism (28). Therefore, our finding of an inverse association of higher blood copper with systolic blood pressure and CAD risk fits with the detrimental effects of copper deficiency.
Strengths and limitations
We used genetic instruments that showed reliable associations in at least two of the three GWAS datasets and showed sufficient strength (>10) with F-statistics of 73.5 for rs17564336 and 37.8 for rs34951015 (relevance assumption). However, as we had only two instruments for the analysis, sensitivity analyses such as MR-Egger that can be used to assess directional horizontal pleiotropy could not be applied. Hence, we cannot exclude potential horizontal pleiotropy by other pathways that were not considered in the MR analyses (exclusion restriction assumption). Database look-ups did not reveal genome-wide significant associations between the two instruments and sociodemographic or lifestyle factors that might confound the copper outcome association (22,29) (independence assumption).
By pooling three different GWAS on copper levels involving up to 6937 individuals to select genetic instruments, we generated the largest genetic data source for blood copper values to date. However, different sample materials were used for copper measurements (serum, whole blood, erythrocytes) and varying transformations of copper values and model adjustments were conducted in the individual GWAS. We tried to overcome limitations due to different analytical approaches by applying the Stouffer Method for meta-analysis, which is robust to different effect estimate scales as it simply requires P-value, sample size and estimated direction of effect for each study (30). However, due to the use of resulting pooled z-scores, neither the beta estimates for associations with copper nor the effect sizes from the MR analysis have interpretable units (31). Still, the results are of value in terms of effect direction.
Moreover, as the liver is the main homeostatic organ for copper and liver content is not accurately reflected by copper levels in plasma or urine (32), blood levels of copper might only weakly correlate with available copper in the organism. Still, for ethical and practical reasons, we cannot use biopsies to determine liver copper content in epidemiological research. In addition, we quantified the total copper content in the different blood fractions and we cannot distinguish between the amount that is bound to Cp and the loosely bound or free copper content.
Our study might be underpowered to detect causal effects for other cardiometabolic risk factors besides systolic blood pressure, where we had a power of 71% (33). Furthermore, we tested multiple phenotypes and the associations for CAD and systolic blood pressure were only nominally significant (<0.05). Using a strict cut-off for multiple testing of 0.017 (0.05/3) or 0.007 (0.05/7) would result in no significant association for disease endpoints or related risk factors, respectively. Although those findings may represent false positives, the result of copper and CAD is in accordance with the previous MR study (6), reinforcing each other. Furthermore, we used the advantage of multivariable MR to study copper and systolic blood pressure in one model. However, we recognized an imbalance of suitable genetic instruments between both exposures of one for copper versus 415 for systolic blood pressure, resulting in a low conditional F-statistic for copper. Although we corrected for weak instruments and received comparable estimates, this analysis should be interpreted cautiously. Furthermore, the outcome summary statistics for blood pressure traits are adjusted for BMI, which might have biased the results due to unmeasured confounding (34). However, these summary statistics represent the largest datasets for blood pressure traits (35) and are not available without BMI adjustment, hence the relationship between copper, blood pressure and CAD needs further investigation. Finally, our findings are restricted to mainly Europeans, and therefore, cannot be generalized to other ethnicities. For CAD, also Asian individuals contributed to the meta-analysis (36), which might be considered as limitation, as individuals included in two-sample MR analyses should come from the same ethnic background. Nevertheless, this should not increase the likelihood of finding an association when there is none (37). Hence, our results on copper and CAD might still be informative in terms of effect direction of causality (38).
Conclusion
In contrast to previous observational evidence establishing higher blood copper levels as risk-increasing factor for cardiometabolic diseases, this study suggests that genetically higher blood copper might play a protective role for CAD development and systolic blood pressure. Whether systolic blood pressure mediates the association between copper and CAD needs further investigation.
Materials and Methods
Study populations
EPIC-Potsdam study
The EPIC-Potsdam study consists of 27 548 participants recruited between 1994 and 1998 from the general population in Potsdam and surroundings, Germany. The baseline examination involved a personal interview, interviewer-conducted anthropometric measurements and a blood sample collection (39). We used a random sample within the EPIC-Potsdam study, described in detail previously (40). Briefly, a sub-cohort of 2500 individuals was randomly selected from 26 444 participants who provided blood samples at baseline. Of these, participants with prevalent diabetes, myocardial infarction or stroke were excluded. Further exclusion criteria were missing genetic data and missing data on copper measurements, leaving 2045 individuals for analyses in the sub-cohort (Supplementary Material, Fig. S4).
All participants provided written informed consent. The EPIC-Potsdam study was approved by the ethics committee of the State of Brandenburg, Germany. All procedures were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
PIVUS study
The PIVUS study aims to investigate the predictive power of different measurements of vascular characteristics for future cardiovascular events (41). Briefly, individuals at the age of 70 years living in the community of Uppsala, Sweden were randomly selected from the community register and invited to participate in the study. Of 2025 invited, 1016 participants were recruited. For the present analysis, GWAS data of 946 individuals were used. The PIVUS study was approved by the Ethical Committee of Uppsala University and each participant gave written informed consent.
QIMR studies
Evans et al. investigated the association between genetic variants and copper levels from erythrocytes in adult twins from different studies conducted at the QIMR, Australia (7). For the present analysis, we utilized updated GWAS data from 3946 individuals. Participants in the QIMR studies gave written informed consent, and the studies were approved by the Human Research Ethics Committee of the QIMR.
Summary-level data for cardiometabolic outcomes
All data resources for cardiometabolic outcomes and cardiometabolic risk factors include individuals with European ancestry and are summarized in Supplementary Material, Table S6. In brief, genetic associations with diabetes were obtained from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium including 74 124 T2D cases and 824 006 controls without BMI adjustment (48.0% women; mean age 59.4 years) (42). Genetic associations with CAD came from the meta-analysis of UK Biobank SOFT CAD GWAS with Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics (CARDIoGRAMplusC4D) 1000 Genomes-based GWAS and the Myocardial Infarction Genetics and CARDIoGRAM Exome (36). The SOFT CAD phenotype incorporates self-reported angina or other evidence of chronic coronary heart disease and 10 801 cases and 137 371 controls (36).
Associations with stroke were obtained from MEGASTROKE (cases = 40 585, controls = 406 111) (43). For the risk factors BMI, high-density lipoprotein (HDL), LDL and blood glucose, we used UK Biobank data (up to 361 194 individuals; 461 460 for BMI) (44). For blood pressure phenotypes, we used GWAS data from the International Consortium of Blood Pressure (mean age ~ 54.5 years, ~ 51% women), UK Biobank and UK BiLEVE (mean age: 56.7 years, 54% women) with up to 757 601 individuals (35). For HbA1c, we used data provided by the MAGIC consortium including 46 368 non-diabetic individuals (52% women) (45).
DNA-extraction, genotyping and quality control within EPIC-Potsdam, PIVUS and QIMR
In EPIC-Potsdam, DNA was extracted from buffy coats using the chemagic DNA Buffy Coat Kit on a Chemagic Magnetic Separation Module I (PerkinElmer chemagen Technologies, Baesweiler, Germany) according to the manufacturer’s manual. Samples were genotyped with three different genotyping arrays: Human660W-Quad_v1_A (n = 363), HumanCoreExome-12v1-0_B (n = 675, two datasets) and Illumina InfiniumOmniExpressExome-8v1-3_A DNA Analysis BeadChip (n = 1377). Genotyping and quality control of the Human660W-Quad_v1_A and HumanCoreExome-12v1-0_B chips was described elsewhere (46). Genotyping using the Illumina InfiniumOmniExpressExome-8v1-3_A DNA Analysis BeadChip was performed in the Life and Brain Center in Bonn, Germany. A detailed description of genotyping, quality control and imputation was previously published (47). Briefly, pre−/phasing and imputation were conducted using Eagle2 (48) and the Michigan Imputation Service (49) with The Haplotype Reference Consortium (release 1.1) as reference panel (50). Imputation was conducted in four separate datasets (one for each chip or two for the HumanCoreExome-12v1-0_B chip) using minimac3 (49). Imputed files were merged, keeping the minimal R2 score from the four files. SNPs were filtered by R2 keeping those with values bigger than 0.6. Overall, we had genotype data for n = 2253 samples from the sub-cohort available (Supplementary Material, Fig. S1).
Genotyping of PIVUS samples was previously described (8). Briefly, samples were genotyped with Illumina Infinium Omni Express bead microarray and imputed according to the Haplotype Reference Consortium reference panel.
QIMR genotypes were determined using Illumina 317, 370 or 610 K chips. Quality control steps and imputation of untyped HapMap 2 SNPs was previously described (51). For the present analysis, we used updated genetic data imputed to 1000 Genomes reference panel.
Blood collection and serum copper measurements in EPIC-Potsdam, PIVUS and QIMR
In EPIC-Potsdam, the method published by Kopp et al. (52) was employed (4) for trace element profiling. Detailed information can be found in the online supplement. In brief, 50 μl of serum sample was diluted with 440 μl of a diluent solution as described in (52). The analysis was conducted via inductively coupled plasma tandem mass spectrometry (ICP-MS/MS) (Agilent ICP-QQQMS 8800, Agilent Technologies, Waldbronn, Germany).
In PIVUS, copper levels were determined in whole blood. The analysis was performed using inductively coupled plasma sector field mass spectrometry, after microwave-assisted digestion with nitric acid (53).
In QIMR, copper levels were measured in erythrocytes (7). Briefly, the erythrocytes were thawed at room temperature and diluted 1:20 in ammonia/ethylenediaminetetraacetic acid (EDTA) solution containing rhodium as an internal standard. Copper concentrations were measured by ICP-MS on a Perkin-Elmer Elan 5000 mass spectrometer (PerkinElmer, Inc., Wellesley, MA, USA) or a Varian UltraMass (Varian Inc., Palo Alto, CA, USA) (7).
Statistical analysis
In EPIC-Potsdam, Statistical Analysis System (SAS) Enterprise Guide 7.1 with SAS version 9.4 (SAS Institute Inc., Cary, NC, USA) was used for data management and preparation. QCtool v1.4 was used for genetic data filtering. SNP test v2.5.2 (54) was used for GWAS.
For meta-analysis of GWAS results, we used METAL (55). For MR analyses we used R (version 3.6.3 (2020-02-29)) and the TwoSampleMR (v0.5.5) (38) and MVMR (v0.2) (56) R packages.
Genome-wide association study in EPIC-Potsdam, PIVUS and QIMR
In EPIC-Potsdam, SNPs were filtered by SNP missing-rate (removed ≥0.05), minimum allele frequency (removed out of interval [0.05–0.5]) and Hardy–Weinberg equilibrium (removed -log10(P-value) ≥3). We performed exploratory single variant association analysis using n = 5 339 559 markers as exposures. We considered a P-value as genome-wide significant at P < 5 × 10−8. Copper levels were natural log-transformed to normalize the right-skewed distributions and standardized. We assumed an additive genetic model, adjusted for age at recruitment and sex. Variants were mapped to Ensembl annotation version 104 (GRCh37) (11,57).
In the PIVUS study, we performed GWAS by applying additive genetic models using inverse rank normalized copper levels and adjusted for sex, cholesterol, triglycerides and two principal components from multi-dimensional scaling of the genetic relationship matrix to account for population structure (8).
In QIMR data, copper levels were log-transformed and adjusted for analysis batch, hemoglobin concentration in the thawed sample and analytical quality control data to generated standardized residuals for subsequent GWAS (7). GWAS on standardized residuals were adjusted for sex and age, using an additive model accounting for within family relatedness (7).
Meta-analysis of GWAS results and instrument selection
We performed meta-analysis of the three different GWAS datasets by applying the Stouffer Method (30) in METAL (55) because different approaches for copper measurement and analysis were used across studies, and effect sizes were therefore not on the same scale. Results were filtered by keeping only SNPs that were available in at least two of the three studies. Based on those, we selected genome-wide significant (P < 5 × 10−8) instrumental variables and performed clumping according to linkage disequilibrium (LD). Therefore, SNPs within a window of 10 000 kb and being in LD as defined by r ≥ 0.001 were removed. The SNPs with the lowest P-values were retained.
Two-sample MR analyses using on copper levels and cardiometabolic outcomes
We conducted a univariable two-sample MR study using copper levels as exposures on cardiometabolic diseases and risk factors. Effect estimates of the association between genetic instruments and copper were obtained from z-scores (31) from meta-analysis and effect estimates of the SNP-outcome association were used from public summary GWAS data (Supplementary Material, Table S6) (35,36,42–45). Data were harmonized for the direction of effects between exposure and outcome associations. We used an inverse variance weighted (IVW) meta-analysis of each SNP specific Wald ratio (SNP-outcome estimate divided by SNP-exposure estimate) using fixed effects and random effects models (the latter as sensitivity analysis). Heterogeneity was assessed by the Cochran’s Q statistic. We obtained the F-statistic as indicator of instrument strength (58).
To assess whether systolic blood pressure might mediate the association between copper and CAD, we conducted multivariable MR (59). Genetic instruments for each exposure were selected based on strict genome-wide significance threshold of 5 × 10−8 and default clumping threshold of r ≥ 0.001 for a combined list of instruments. Then we obtained all SNP effects on the respective other exposure. The extraction of the outcome GWAS results and the data harmonization was conducted as described before. We calculated conditional F-statistics to evaluate the presence of weak instruments and adjusted for those by minimizing the Q-statistic allowing for heterogeneity using ‘qhet_mvmr’ function from the MVMR package (56). To assess the correlation between systolic blood pressure and copper we used EPIC-Potsdam data. 95% CIs for the effect estimates were determined using non-parametric bootstrap with 1000 iterations.
Data Availability
The EPIC-Potsdam datasets analyzed during the current study are not publicly available due to data protection regulations. In accordance with German Federal and State data protection regulations, epidemiological data analyses of EPIC-Potsdam may be initiated upon an informal inquiry addressed to the secretariate of the Human Study Center (Office.HSZ@dife.de). Each request will then have to pass a formal process of application and review by the respective PI and a scientific board.
Summary-statistics of this meta-analysis of GWAS on blood copper values are available through the GWAS catalog.
Summary-level data for genetic associations with type 2 diabetes, CAD and stroke have been contributed by the DIAGRAM consortium (http://diagram-consortium.org/downloads.html; accessed 13.08.2019), the Cardiogram consortium (http://www.cardiogramplusc4d.org/; accessed 12.08.2019) and MEGASTROKE (https://www.megastroke.org/; accessed 05.02.2020). The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. GWAS-Summary data from cardiometabolic risk factors were accessed via the MR-Base platform and TwoSampleMR R package (38). The authors thank all investigators for sharing these data.
Supplementary Material
Acknowledgements
We thank the Human Study Centre (HSC) of the German Institute of Human Nutrition Potsdam-Rehbrücke, namely the trustee and the data hub for the processing, and the participants for the provision of the data, the biobank for the processing of the biological samples and the head of the HSC, Manuela Bergmann, for the contribution to the study design and leading the underlying processes of data generation. Conflict of Interest statement. None declared.
Contributor Information
Susanne Jäger, Department of Molecular Epidemiology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal 14558, Germany; German Center for Diabetes Research (DZD), Neuherberg 85764, Germany; TraceAge-DFG Research Unit on Interactions of Essential Trace Elements in Healthy and Diseased Elderly, Potsdam-Berlin-Jena, Germany.
Maria Cabral, Department of Molecular Epidemiology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal 14558, Germany; German Center for Diabetes Research (DZD), Neuherberg 85764, Germany; TraceAge-DFG Research Unit on Interactions of Essential Trace Elements in Healthy and Diseased Elderly, Potsdam-Berlin-Jena, Germany.
Johannes F Kopp, TraceAge-DFG Research Unit on Interactions of Essential Trace Elements in Healthy and Diseased Elderly, Potsdam-Berlin-Jena, Germany; Department of Food Chemistry, Institute of Nutritional Science, University of Potsdam, Nuthetal 14558, Germany.
Per Hoffmann, Human Genomics Research Group, Department of Biomedicine, University of Basel, Basel 4031, Switzerland; Division of Genomics, Life and Brain Research Centre, Institute of Human Genetics, University Hospital of Bonn, Bonn 53105, Germany.
Esther Ng, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK.
John B Whitfield, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4029, Australia.
Andrew P Morris, Centre for Genetics and Genomics Versus Arthritis, Centre for Musculoskeletal Research, The University of Manchester, Manchester M13 9PL, UK.
Lars Lind, Department of Medical Sciences, Cardiovascular Epidemiology, Uppsala University, Uppsala 75185, Sweden.
Tanja Schwerdtle, TraceAge-DFG Research Unit on Interactions of Essential Trace Elements in Healthy and Diseased Elderly, Potsdam-Berlin-Jena, Germany; Department of Food Chemistry, Institute of Nutritional Science, University of Potsdam, Nuthetal 14558, Germany; Department Food Safety, German Federal Institute for Risk Assessment (BfR), Berlin 10589, Germany.
Matthias B Schulze, Department of Molecular Epidemiology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal 14558, Germany; German Center for Diabetes Research (DZD), Neuherberg 85764, Germany; TraceAge-DFG Research Unit on Interactions of Essential Trace Elements in Healthy and Diseased Elderly, Potsdam-Berlin-Jena, Germany; Institute of Nutritional Science, University of Potsdam, Nuthetal 14558, Germany.
Funding
This work was funded by TraceAge—DFG Research Unit on Interactions of essential trace elements in healthy and diseased elderly, Potsdam-Berlin-Jena (FOR 2558/1). The recruitment phase of the EPIC-Potsdam Study was supported by the Federal Ministry of Science, Germany (01 EA 9401) and the European Union (SOC 95201408 05F02). The follow-up of the EPIC-Potsdam Study was supported by German Cancer Aid (70-2488-Ha I) and the European Community (SOC 98200769 05F02). This work was furthermore supported by a grant from the German Ministry of Education and Research (BMBF) and the State of Brandenburg (DZD grant 82DZD00302).
The PIVUS study obtained funding from the Uppsala University Hospital.
Recruitment and interviewing of participants, sample collection, and trace element measurements for the QIMR studies were funded by grants AA007535, AA013320, AA013321, AA013326, AA014041 and DA012854 from the US National Institutes of Health.
Author contributions
S.J. and M.B.S. conceptualized the project and designed the analysis plan. S.J. analysed the data and performed meta-analysis. A.M., E.N. and J.W. conducted GWAS studies. P.H., J.F.K and T.S. acquired data. S.J., M.C., A.M. and M.B.S. interpreted the data. S.J. wrote the first draft of the manuscript. M.C., P.H., J.F.K, E.N., J.W., A.M., L.L., T.S. and M.B.S. reviewed the manuscript. Both S.J. and M.B.S had access to all data for this study and take responsibility for the manuscript contents. All authors approved the final version to be published.
References
- 1. Cabral, M., Kuxhaus, O., Eichelmann, F., Kopp, J.F., Alker, W., Hackler, J., Kipp, A.P., Schwerdtle, T., Haase, H., Schomburg, L.et al. (2021) Trace element profile and incidence of type 2 diabetes, cardiovascular disease and colorectal cancer: results from the EPIC-Potsdam cohort study. Eur. J. Nutr., 60, 3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiao, Y., Yuan, Y., Liu, Y., Yu, Y., Jia, N., Zhou, L., Wang, H., Huang, S., Zhang, Y., Yang, H.et al. (2019) Circulating multiple metals and incident stroke in Chinese adults. Stroke, 50, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Malavolta, M., Piacenza, F., Basso, A., Giacconi, R., Costarelli, L. and Mocchegiani, E. (2015) Serum copper to zinc ratio: relationship with aging and health status. Mech. Ageing Dev., 151, 93–100. [DOI] [PubMed] [Google Scholar]
- 4. Baudry, J., Kopp, J.F., Boeing, H., Kipp, A.P., Schwerdtle, T. and Schulze, M.B. (2020) Changes of trace element status during aging: results of the EPIC-Potsdam cohort study. Eur. J. Nutr., 59, 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davey Smith, G. and Ebrahim, S. (2003) 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol., 32, 1–22. [DOI] [PubMed] [Google Scholar]
- 6. Kodali, H.P., Pavilonis, B.T. and Schooling, C.M. (2018) Effects of copper and zinc on ischemic heart disease and myocardial infarction: a Mendelian randomization study. Am. J. Clin. Nutr., 108, 237–242. [DOI] [PubMed] [Google Scholar]
- 7. Evans, D.M., Zhu, G., Dy, V., Heath, A.C., Madden, P.A., Kemp, J.P., McMahon, G., St Pourcain, B., Timpson, N.J., Golding, J.et al. (2013) Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum. Mol. Genet., 22, 3998–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ng, E., Lind, P.M., Lindgren, C., Ingelsson, E., Mahajan, A., Morris, A. and Lind, L. (2015) Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum. Mol. Genet., 24, 4739–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leone, N., Courbon, D., Ducimetiere, P. and Zureik, M. (2006) Zinc, copper, and magnesium and risks for all-cause, cancer, and cardiovascular mortality. Epidemiology, 17, 308–314. [DOI] [PubMed] [Google Scholar]
- 10. Grammer, T.B., Kleber, M.E., Silbernagel, G., Pilz, S., Scharnagl, H., Lerchbaum, E., Tomaschitz, A., Koenig, W. and März, W. (2014) Copper, ceruloplasmin, and long-term cardiovascular and total mortality (the Ludwigshafen risk and cardiovascular health study). Free Radic. Res., 48, 706–715. [DOI] [PubMed] [Google Scholar]
- 11. Ensembl database . Available fromhttp://grch37.ensembl.org. Accessed 15.06.2021
- 12. Schooling, C.M., Lopez, P.M., Yang, Z., Zhao, J.V., Au Yeung, S.L. and Huang, J.V. (2020) Use of multivariable Mendelian randomization to address biases due to competing risk before recruitment. Front. Genet., 11, 610852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou, J., Liu, C., Francis, M., Sun, Y., Ryu, M.S., Grider, A. and Ye, K. (2020) The causal effects of blood iron and copper on lipid metabolism diseases: evidence from phenome-wide Mendelian randomization study. Nutrients, 12, 3174–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carpenter, W.E., Lam, D., Toney, G.M., Weintraub, N.L. and Qin, Z. (2013) Zinc, copper, and blood pressure: human population studies. Med. Sci. Monit., 19, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adamsson Eryd, S., Sjögren, M., Smith, J.G., Nilsson, P.M., Melander, O., Hedblad, B. and Engström, G. (2014) Ceruloplasmin and atrial fibrillation: evidence of causality from a population-based Mendelian randomization study. J. Intern. Med., 275, 164–171. [DOI] [PubMed] [Google Scholar]
- 16. Reunanen, A., Knekt, P. and Aaran, R.K. (1992) Serum ceruloplasmin level and the risk of myocardial infarction and stroke. Am. J. Epidemiol., 136, 1082–1090. [DOI] [PubMed] [Google Scholar]
- 17. Engström, G., Lind, P., Hedblad, B., Stavenow, L., Janzon, L. and Lindgärde, F. (2002) Effects of cholesterol and inflammation-sensitive plasma proteins on incidence of myocardial infarction and stroke in men. Circulation, 105, 2632–2637. [DOI] [PubMed] [Google Scholar]
- 18. GTEx Consortium (2013) The genotype-tissue expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Braga, C.P., Vieira, J.C.S., Grove, R.A., Boone, C.H.T., Leite, A.L., Buzalaf, M.A.R., Fernandes, A.A.H., Adamec, J. and Padilha, P.M. (2017) A proteomic approach to identify metalloproteins and metal-binding proteins in liver from diabetic rats. Int. J. Biol. Macromol., 96, 817–832. [DOI] [PubMed] [Google Scholar]
- 20. Kühn, E.C., Slagman, A., Kühn-Heid, E.C.D., Seelig, J., Schwiebert, C., Minich, W.B., Stoppe, C., Möckel, M. and Schomburg, L. (2019) Circulating levels of selenium-binding protein 1 (SELENBP1) are associated with risk for major adverse cardiac events and death. J. Trace Elem. Med. Biol., 52, 247–253. [DOI] [PubMed] [Google Scholar]
- 21. Kühn-Heid, E.C.D., Kühn, E.C., Ney, J., Wendt, S., Seelig, J., Schwiebert, C., Minich, W.B., Stoppe, C. and Schomburg, L. (2019) Selenium-binding protein 1 indicates myocardial stress and risk for adverse outcome in cardiac surgery. Nutrients, 11, 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. IEU OpenGWAS Project . Available fromhttps://gwas.mrcieu.ac.uk/phewas/. Accessed 20.8.2021
- 23. Pernow, J., Mahdi, A., Yang, J. and Zhou, Z. (2019) Red blood cell dysfunction: a new player in cardiovascular disease. Cardiovasc. Res., 115, 1596–1605. [DOI] [PubMed] [Google Scholar]
- 24. Holeček, M. (2020) Histidine in health and disease: metabolism, physiological importance, and use as a supplement. Nutrients, 12, 848–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klevay, L.M. (2000) Cardiovascular disease from copper deficiency--a history. J. Nutr., 130, 489s–492s. [DOI] [PubMed] [Google Scholar]
- 26. DiNicolantonio, J.J., Mangan, D. and O'Keefe, J.H. (2018) Copper deficiency may be a leading cause of ischaemic heart disease. Open. Heart., 5, e000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schuschke, D.A. (1997) Dietary copper in the physiology of the microcirculation. J. Nutr., 127, 2274–2281. [DOI] [PubMed] [Google Scholar]
- 28. Burkhead, J.L. and Lutsenko, S. (2013) The role of copper as a modifier of lipid metabolism. In Baez, R.V. (ed), Lipid Metabolism InTech, London, SW7 2QJ, UK. [Google Scholar]
- 29. Phenoscanner . Available fromhttp://www.phenoscanner.medschl.cam.ac.uk/. Accessed 20.8.2021
- 30. Stouffer, S.A., Suchman, E.A., Devinney, L.C., Star, S.A. and Williams, R.M., Jr. (1949) The American Soldier: Adjustment During Army Life. (Studies in Social Psychology in World War II), Vol. 1. Princeton Univ. Press, Oxford, England. [Google Scholar]
- 31. Taylor, A.E., Burgess, S., Ware, J.J., Gage, S.H., Richards, J.B., Davey Smith, G. and Munafò, M.R. (2016) Investigating causality in the association between 25(OH)D and schizophrenia. Sci. Rep., 6, 26496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans, J., Newman, S. and Sherlock, S. (1978) Liver copper levels in intrahepatic cholestasis of childhood. Gastroenterology, 75, 875–878. [PubMed] [Google Scholar]
- 33. Deng, L., Zhang, H. and Yu, K. (2020) Power calculation for the general two-sample Mendelian randomization analysis. Genet. Epidemiol., 44, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hartwig, F.P., Tilling, K., Davey Smith, G., Lawlor, D.A. and Borges, M.C. (2021) Bias in two-sample Mendelian randomization when using heritable covariable-adjusted summary associations. Int. J. Epidemiol., 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evangelou, E., Warren, H.R., Mosen-Ansorena, D., Mifsud, B., Pazoki, R., Gao, H., Ntritsos, G., Dimou, N., Cabrera, C.P., Karaman, I.et al. (2018) Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet., 50, 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson, C.P., Goel, A., Butterworth, A.S., Kanoni, S., Webb, T.R., Marouli, E., Zeng, L., Ntalla, I., Lai, F.Y., Hopewell, J.C.et al. (2017) Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet., 49, 1385–1391. [DOI] [PubMed] [Google Scholar]
- 37. Burgess, S., Butterworth, A.S. and Thompson, J.R. (2016) Beyond Mendelian randomization: how to interpret evidence of shared genetic predictors. J. Clin. Epidemiol., 69, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hemani, G., Zheng, J., Elsworth, B., Wade, K.H., Haberland, V., Baird, D., Laurin, C., Burgess, S., Bowden, J., Langdon, R.et al. (2018) The MR-Base platform supports systematic causal inference across the human phenome. elife, 7, e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boeing, H., Wahrendorf, J. and Becker, N. (1999) EPIC-Germany--A source for studies into diet and risk of chronic diseases. European investigation into cancer and nutrition. Ann. Nutr. Metab., 43, 195–204. [DOI] [PubMed] [Google Scholar]
- 40. Stefan, N., Fritsche, A., Weikert, C., Boeing, H., Joost, H.G., Haring, H.U. and Schulze, M.B. (2008) Plasma Fetuin-a levels and the risk of type 2 diabetes. Diabetes, 57, 2762–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lind, L., Fors, N., Hall, J., Marttala, K. and Stenborg, A. (2005) A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the prospective investigation of the vasculature in Uppsala seniors (PIVUS) study. Arterioscler. Thromb. Vasc. Biol., 25, 2368–2375. [DOI] [PubMed] [Google Scholar]
- 42. Mahajan, A., Taliun, D., Thurner, M., Robertson, N.R., Torres, J.M., Rayner, N.W., Payne, A.J., Steinthorsdottir, V., Scott, R.A., Grarup, N.et al. (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet., 50, 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malik, R., Chauhan, G., Traylor, M., Sargurupremraj, M., Okada, Y., Mishra, A., Rutten-Jacobs, L., Giese, A.K., van derLaan, S.W., Gretarsdottir, S.et al. (2018) Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet., 50, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UK Biobank GWAS 2018. Available fromhttp://www.nealelab.is/uk-biobank/. Accessed 11.6.2021 via MR-Base
- 45. Soranzo, N., Sanna, S., Wheeler, E., Gieger, C., Radke, D., Dupuis, J., Bouatia-Naji, N., Langenberg, C., Prokopenko, I., Stolerman, E.et al. (2010) Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes, 59, 3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langenberg, C., Sharp, S.J., Franks, P.W., Scott, R.A., Deloukas, P., Forouhi, N.G., Froguel, P., Groop, L.C., Hansen, T., Palla, L.et al. (2014) Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med., 11, e1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jäger, S., Cuadrat, R., Hoffmann, P., Wittenbecher, C. and Schulze, M.B. (2020) Desaturase activity and the risk of type 2 diabetes and coronary artery disease: a Mendelian randomization study. Nutrients, 12, 2261–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loh, P.R., Palamara, P.F. and Price, A.L. (2016) Fast and accurate long-range phasing in a UK biobank cohort. Nat. Genet., 48, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Das, S., Forer, L., Schonherr, S., Sidore, C., Locke, A.E., Kwong, A., Vrieze, S.I., Chew, E.Y., Levy, S., McGue, M.et al. (2016) Next-generation genotype imputation service and methods. Nat. Genet., 48, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCarthy, S., Das, S., Kretzschmar, W., Delaneau, O., Wood, A.R., Teumer, A., Kang, H.M., Fuchsberger, C., Danecek, P., Sharp, K.et al. (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet., 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Medland, S.E., Nyholt, D.R., Painter, J.N., McEvoy, B.P., McRae, A.F., Zhu, G., Gordon, S.D., Ferreira, M.A., Wright, M.J., Henders, A.K.et al. (2009) Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet., 85, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kopp, J.F., Müller, S.M., Pohl, G., Lossow, K., Kipp, A.P. and Schwerdtle, T. (2019) A quick and simple method for the determination of six trace elements in mammalian serum samples using ICP-MS/MS. J. Trace Elem. Med. Biol., 54, 221–225. [DOI] [PubMed] [Google Scholar]
- 53. Rodushkin, I., Ödman, F., Olofsson, R. and Axelsson, M.D. (2000) Determination of 60 elements in whole blood by sector field inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom., 15, 937–944. [Google Scholar]
- 54. Marchini, J., Howie, B., Myers, S., McVean, G. and Donnelly, P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. [DOI] [PubMed] [Google Scholar]
- 55. Willer, C.J., Li, Y. and Abecasis, G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanderson, E., Spiller, W. and Bowden, J. (2021) Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomisation. Stat. Med., 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cunningham, F., Amode, M.R., Barrell, D., Beal, K., Billis, K., Brent, S., Carvalho-Silva, D., Clapham, P., Coates, G., Fitzgerald, S.et al. (2015) Ensembl 2015. Nucleic Acids Res., 43, D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Palmer, T.M., Lawlor, D.A., Harbord, R.M., Sheehan, N.A., Tobias, J.H., Timpson, N.J., Davey Smith, G. and Sterne, J.A. (2012) Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res., 21, 223–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burgess, S. and Thompson, S.G. (2015) Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol., 181, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EPIC-Potsdam datasets analyzed during the current study are not publicly available due to data protection regulations. In accordance with German Federal and State data protection regulations, epidemiological data analyses of EPIC-Potsdam may be initiated upon an informal inquiry addressed to the secretariate of the Human Study Center (Office.HSZ@dife.de). Each request will then have to pass a formal process of application and review by the respective PI and a scientific board.
Summary-statistics of this meta-analysis of GWAS on blood copper values are available through the GWAS catalog.
Summary-level data for genetic associations with type 2 diabetes, CAD and stroke have been contributed by the DIAGRAM consortium (http://diagram-consortium.org/downloads.html; accessed 13.08.2019), the Cardiogram consortium (http://www.cardiogramplusc4d.org/; accessed 12.08.2019) and MEGASTROKE (https://www.megastroke.org/; accessed 05.02.2020). The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. GWAS-Summary data from cardiometabolic risk factors were accessed via the MR-Base platform and TwoSampleMR R package (38). The authors thank all investigators for sharing these data.


