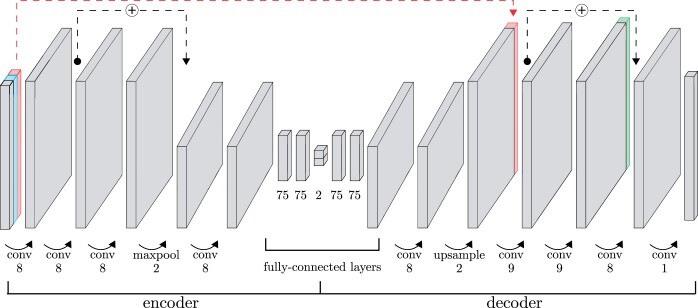
Fig. 1.
Architecture of the GCAE model used for dimensionality reduction. The encoder transforms data to a lower-dimensional latent representation through a series of convolutional, pooling and fully-connected layers. The decoder reconstructs the input genotypes. The input consists of three layers: genotype data (gray), a binary mask representing missing data (blue), and a marker-specific trainable variable per SNP (red). The red dashed line indicates where this marker-specific variable is concatenated to a layer in the decoder. Another marker-specific trainable variable, shown in green, is also concatenated to the second-last layer in the decoder. Black dashed lines indicate residual connections, where the output of a layer is added to that of another layer later in the network. The numbers below the layers indicate the number of kernels for convolutional layers, down- or upsampling factor for pooling and upsampling layers, and number of units for fully-connected layers. The displayed numbers are those of the final model used to obtain the presented results for dimensionality reduction to two dimensions. For other numbers of dimensions, the only modification made was to change the number of units in the latent representation from 2 to 4, 6, 8, or 10. For the genetic clustering application, the number of units in the latent representation was k = 5.