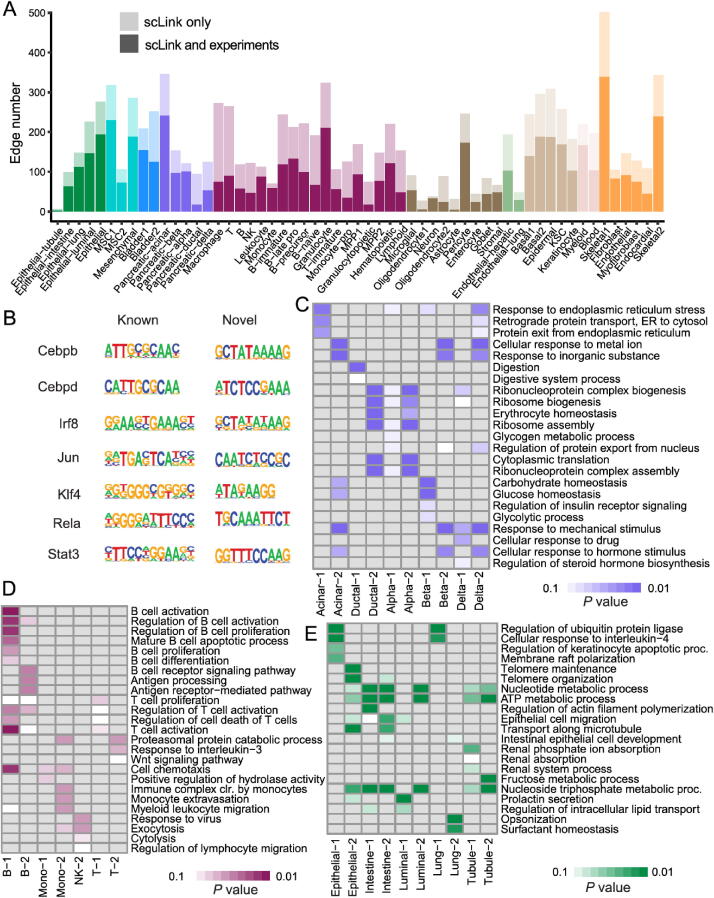
Figure 3.
Performance of scLink on the Tabula Muris dataset. A. The numbers of TF–gene edges identified only by scLink and by both scLink and ChIP-seq experiments. B. Known and novel motifs of seven TFs identified from the promoter regions of genes connected to these TFs by scLink. C. Enriched GO terms in the two largest gene modules identified from pancreatic cell types. D. Enriched GO terms in the two largest gene modules identified from immune cell types. E. Enriched GO terms in the two largest gene modules identified from epithelial cell types. FDR-adjusted P values are shown in the heatmaps, and P values greater than 0.1 are shown in gray. TF, transcription factor; FDR, false discovery rate.