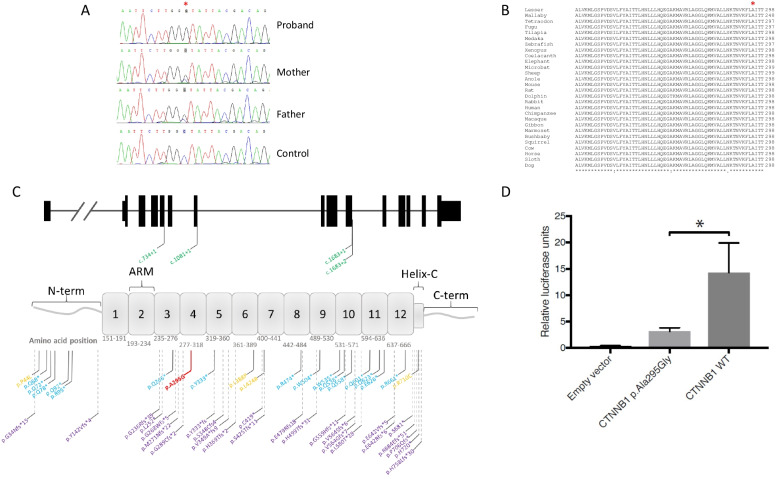
Fig. 1.
Localisation, conservation and transcriptional activity of CTNNB1 c.884C>G; p.(Ala295Gly). A Sanger sequencing chromatograms confirming the presence and zygosity of the CTNNB1 c.884C>G; p.(Ala295Gly) variant in proband and parental DNA samples, in comparison to a control chromatogram which does not contain the sequence change. B CTNNB1 (NM_001904) gene (top) and β-catenin protein (bottom) schematics annotated with previously reported dominant disease-causing mutations (as reported by HGMDpro; orange text = missense mutations, green text = splice-altering mutations; blue text = nonsense mutations; purple text = small insertion, deletion or insertion/deletion mutations) and the location of the CTNNB1 c.884C>G; p.(Ala295Gly) homozygous variant identified by this study (indicated by red text). The biallelic missense mutation identified herein resides within the fourth armadillo (ARM) domain of the encoded protein. C Multispecies alignment of β-catenin (human amino acids 239–298) showing 100% conservation across 28 species of the alanine residue at position 295, found to be mutated to Glycine in the proband described in this report. Red asterisk (*) above the alignments indicates the position of human Alanine 295 in the alignment. Amino acid residues that are fully conserved are represented by an asterisk underneath the alignments; conservation between residues with strongly similar properties are represented by a colon (:); conservation between residues with weakly similar properties are represented by a period symbol (.). D Functional assessment of p.(Ala295Gly) mutant β-catenin on transcription. Bar chart detailing TOPflash luciferase assay results of STF cells transiently co-transfected with wildtype (WT) or mutant (p.(Ala295Gly)) β-catenin or empty vector (pDEST40) and renilla luciferase plasmid. Luciferase activity was measure 48 h post-transfection. Each bar indicates the recorded luciferase activity for each construct following normalisation to renilla activity and relative to the measurement recorded for cells transfected with empty vector. Results are from three independent experiments performed in triplicate. Error bars denote standard error of the mean (SEM); Y-axis represents the fold difference relative to the observed empty vector reading