Figure 5. Protein engagement profiles for Palmostatin M as determined by MS-ABPP using the Palm M-yne probe.
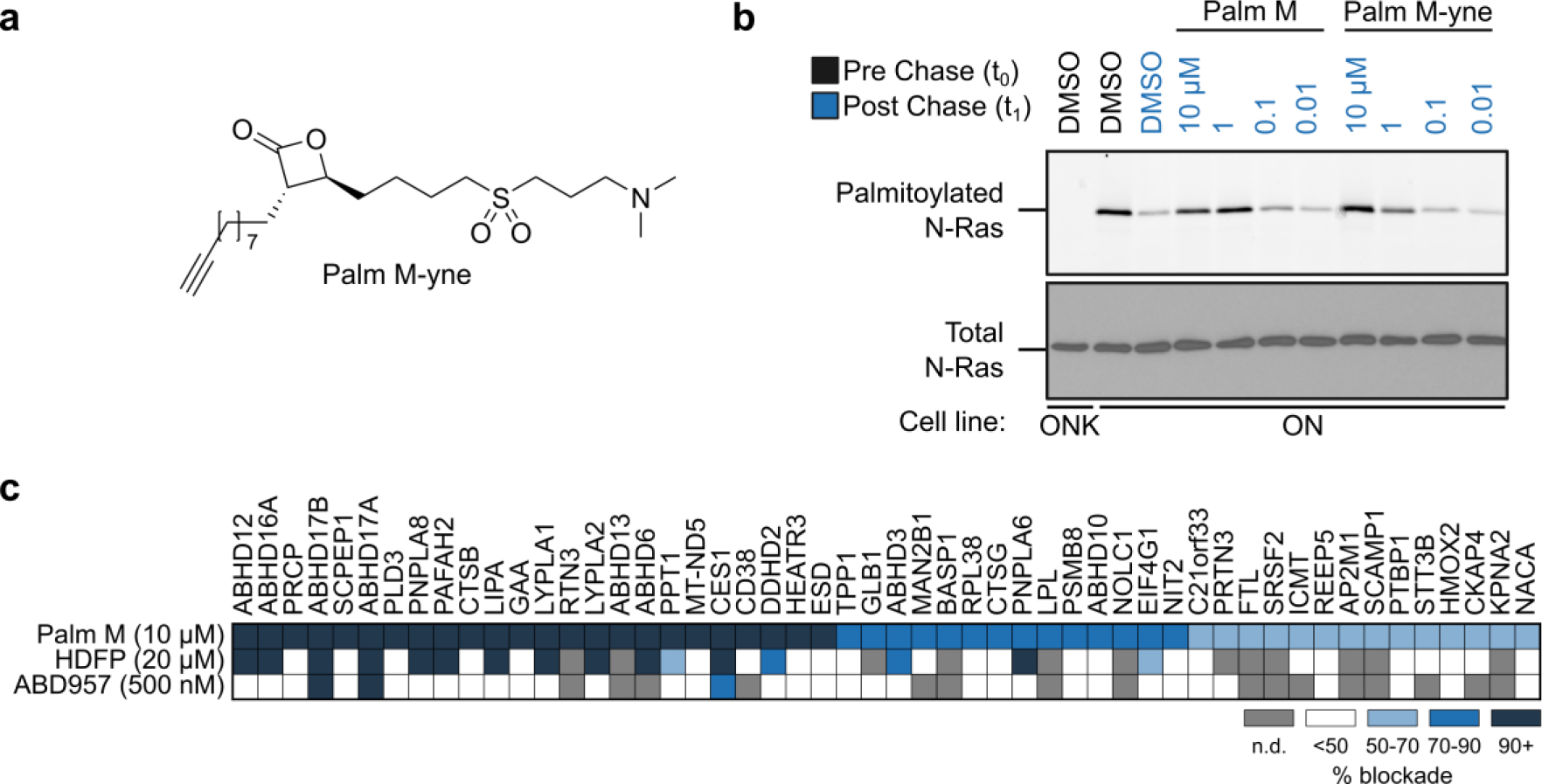
a, Structure of the Palm M-yne probe9. b, Effect of Palm M-yne probe on N-Ras palmitoylation as detected by pulse-chase metabolic labeling of ON cells (or ONK control cells) with the 17-ODYA probe. N-Ras was immunoprecipitated using anti-GFP antibodies prior to visualizing palmitoylation by CuAAC conjugation to a Rh-N3 reporter tag (top panel). Total N-Ras content was measured by Western blotting of GFP enrichments (bottom panel). The concentration-dependent analysis of Palm M and Palm M-yne blockade of N-Ras depalmitoylation was performed once. c, Heatmap summarizing results from competitive MS-ABPP experiments with Palm M (10 μM; n = 3), HDFP (20 μM; n = 2), and ABD957 (500 nM; n = 1) in ON cells treated in situ for 2 h prior to lysis, fractionation, and treatment of the particulate proteome with Palm M-yne (10 μM, 1 h). Values plotted represent percent of blockade of Palm M-yne enrichment by competitor ligands for each protein compared to DMSO control. Heatmap shows only targets that were enriched > 5-fold by the Palm M-yne probe (compared to DMSO control for enrichment; n = 2) and showing > 50% blockade of enrichment by Palm M (compared to DMSO control for competition). For full dataset, see Supp. Table 1. Grey color marks proteins that were not detected or failed to pass quality filters in the indicated experiments. Insufficient numbers of unique peptides were obtained to quantify ABHD17C in this experiment. MS-ABPP data represent average values from independent experiments, as indicated for each condition.
