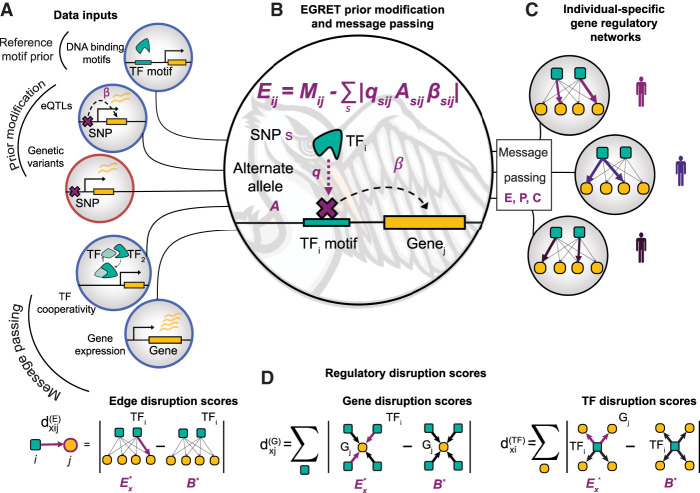
Figure 1.
EGRET integrates multiple data types to construct individual-specific GRNs. (A) EGRET takes as input several data types (population-level inputs circled in blue, individual-specific inputs circled in orange) to construct individual-specific GRNs: an initial estimate of the binding locations of TFs in the form of a reference motif prior (Mij), the beta values of eQTL associations between “eSNPs” and “eGenes” (β), the genetic variants (s) harbored by the individual in question, PPI data as an estimate of TF–TF cooperativity (P), and gene expression to estimate a gene coexpression matrix (C). (B) An individual's genetic variants are used to modify the reference motif prior to produce an individual-specific EGRET prior (E) by penalizing motif-gene connections in which that individual carries a variant allele (A) in the relevant promoter-region motif such that the variant is an eQTL for the adjacent gene (β) and the variant is predicted by QBiC to affect TF binding at that location (q). (C) Message passing is used to integrate the coexpression (C) and PPI (P) networks with the EGRET prior (E) resulting in a final, unique GRN per individual (E*). (D) Regulatory disruption scores can be calculated to quantify the extent to which an edge or node in the network is disrupted by variants. Edge disruption scores are calculated by subtracting a genotype-agnostic baseline network (B*) from the individual's EGRET network and taking the absolute value. TF or gene disruption scores are calculated taking the sum of the edge disruption scores around the TF or gene in question.