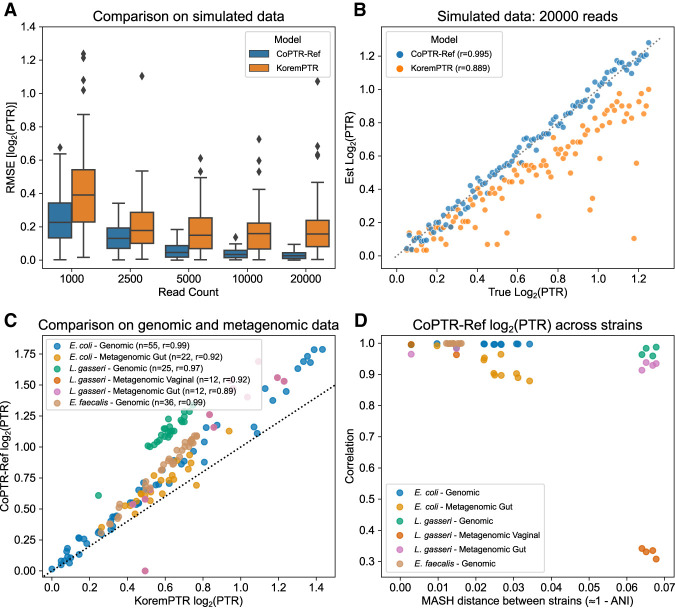
Figure 2.
CoPTR-Ref is accurate on simulated and real data. (A) Accuracy of CoPTR-Ref and KoremPTR on simulated data based on an E. coli genome. Performance was compared by computing the root-mean-square-error (RMSE) of the log2(PTR) (y-axis) across 100 replicates while varying the number of reads (x-axis), varying the position of the replication origin, and varying the PTR. (B) Ground truth (x-axis) and estimated (y-axis) log2(PTR) across 100 simulation replicates with 20,000 reads. KoremPTR appears to underestimate the true log2(PTR). (C) Comparison of KoremPTR log2(PTR) (x-axis) and CoPTR log2(PTR) (y-axis) on six real genomic and metagenomic data sets. (D) Evaluation of CoPTR-Ref's log2(PTR) estimates using representative genomes from different strains (five E. coli strains, four L. gasseri strains, and five E. faecalis strains). Each data set in panel C was mapped to strains from the same species, and the Pearson's correlation (y-axis) was computed for each pair of strains. When the distance between strains (x-axis) is small, log2(PTR)s are highly correlated.