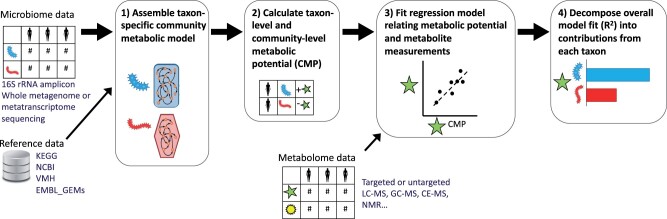
Fig. 1.
Summary of the MIMOSA2 analysis pipeline. In a MIMOSA2 analysis, microbiome data features are first linked to pre-processed reference databases to construct a community metabolic model describing the predicted metabolic reaction capabilities of each community member taxon (Step 1). Next, this network model is combined with microbiome feature abundances to calculate CMP scores for each metabolite, taxon and sample, representing the approximate relative capacity to synthesize or utilize that metabolite (Step 2). Total CMP scores for each metabolite are linked to metabolomics measurements using a regression model (Step 3). For metabolites with a significant relationship between concentration and potential, the specific taxonomic contributors to each metabolite are then analyzed (Step 4)