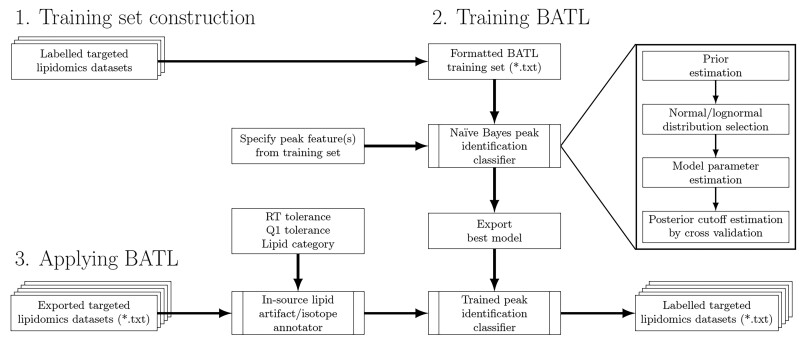
Fig. 2.
Schematic of the BATL lipid identification workflow. BATL follows three steps: (i) users are asked to identify training datasets for which they have unambiguous knowledge of peak identities. (ii) These datasets are used to train BATL, constructing a naïve Bayes statistical model based on the peak features users select. (iii) The model and associated metadata are used by the BATL algorithm to annotate peaks in subsequent query SRM, MRM or PRM datasets