Fig. 1.
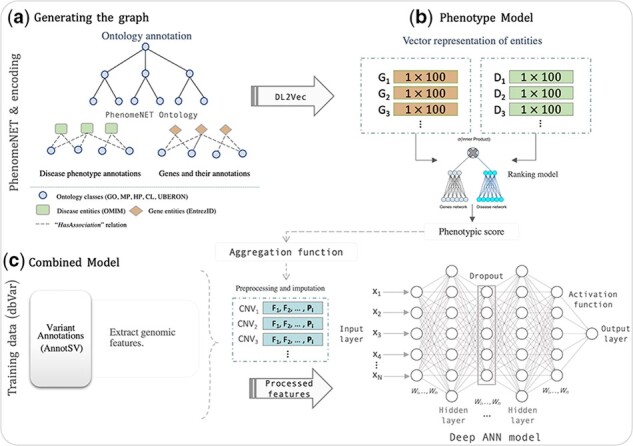
Overview over the DeepSVP model. (a) First, a graph is generated from the ontology axioms in which nodes represent classes or entities annotated with ontology classes, and edges represent axioms that hold between these classes. (b) The DL2Vec workflow takes a set of phenotypes as input and predicts whether a gene is likely associated with these phenotypes using the background knowledge in the graph generated from the ontology and its associations. (c) The combined model uses the prediction score of the DL2Vec phenotype model combined with genomic features derived from SVs. The model outputs a prediction score for each variant that determines how likely the variant is causative of the phenotypes provided as input. G, genes; D, diseases; F, (genomic) features; P, phenotypic score
