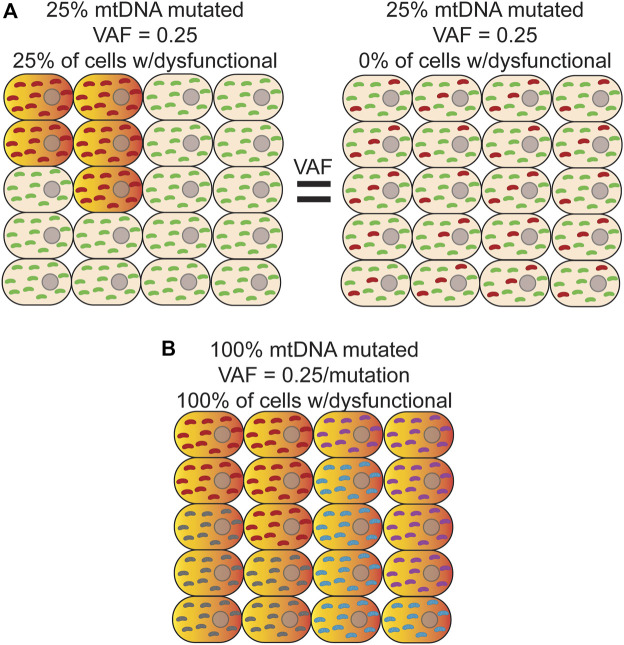
FIGURE 2.
Bulk analysis of mtDNA mutations is unable to distinguish different scenarios with different physiological consequences. (A) Bulk sequencing of cells where left-25% of cells harbor a homoplasmic mutation (red) that causes OXPHOS dysfunction (orange/red-gradient) and the remainder contained only wild-type mtDNA (green) would have the same variant allele fraction (VAF) as bulk sequencing of a sample where Right-100% of cells contain a mutation that is, on average, at 25% heteroplasmy. In both cases, the apparent VAF would be 0.25, but the effect on tissue function would be different. Distinguishing between these two scenarios is only possible with single cell sequencing. (B) In this example, every cell is a clonal expansion harboring one of five possible homoplasmic mtDNA mutations (red, purple, grey, and cyan) that results in OXPHOS dysfunction. In bulk sequencing, no mutation would be seen to have a VAF 0.25, which is below the apparent phenotypic threshold.