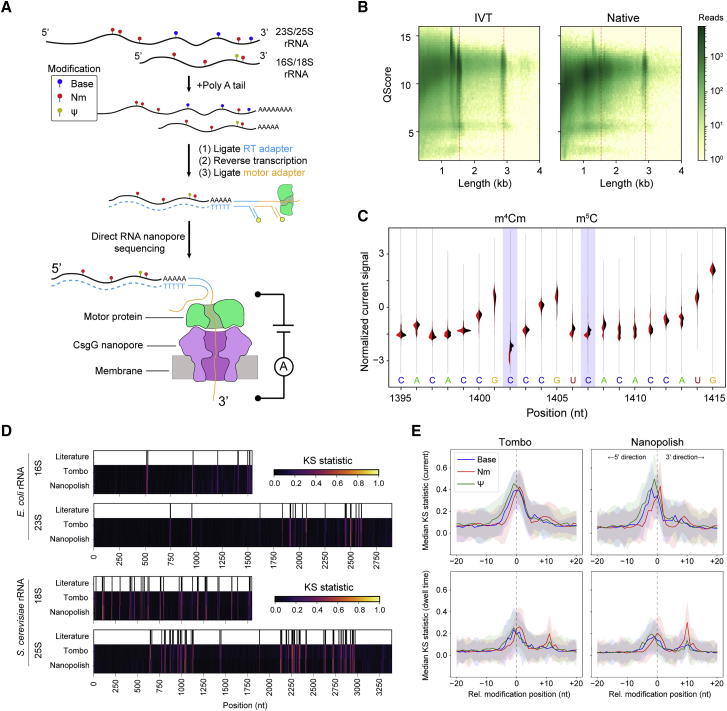
Figure 1.
Direct RNA nanopore sequencing and modification detection
(A) Scheme for direct RNA nanopore sequencing. rRNAs containing native modifications are poly(A) tailed before ligation of adapters and RT. Ionic current blockage events are characteristic of the kmer sequence of RNA transiting through the pore constriction.
(B) Read quality heatmap for IVT rRNA (left) and native rRNA (right) from E. coli. Dashed red lines indicated the expected lengths for 16S rRNA (1.5 kb) and 23S rRNA (2.9 kb)
(C) Normalized native (red) and IVT (black) current signal alignment for 16S rRNA from E. coli spanning positions 1395–1415 performed using Tombo. Sites of known modifications within this window are highlighted in blue.
(D) Positional Kolmogorov-Smirnov (KS) statistical testing of current signals across rRNA from E. coli and S. cerevisiae using both Tombo and Nanopolish. Modification positions described in the literature are indicated as black lines.
(E) Median current and dwell KS statistic profiles separated by modification type (base [excluding ], blue; 2′-O-methyl, red; and , green) and aligned by modification position from both Tombo and Nanopolish. Colored, shaded regions represent the standard deviation of the KS statistic.