Abstract
SARS-CoV-2 has kept the world in suspense for almost 2 years now. The virus spread rapidly worldwide and several variants of concern have emerged: Alpha, Beta, Gamma, Delta and recently Omicron. A rapid method to detect key mutations is needed because these variants may jeopardize the effectiveness of immune protection following vaccination or past infection. This article describes an easy, cheap and fast method for the detection of mutations in the spike protein that are indicative for specific variants. This method can easily distinguish Omicron from other variants.
Keywords: SARS-CoV-2 Omicron, Easy detection of variants of concern, RT-PCR, Sanger sequencing
Since autumn 2020, various mutations that affect the characteristics of SARS-CoV-2 have been reported. Important mutations that occur in the S protein result in the emergence and sustained transmission of multiple variants of concern (VOC) (Zhang et al., 2020). These VOCs are associated with increased transmissibility and are defined by multiple convergent mutations that are hypothesized to have arisen in the context of chronic infections or in previously infected individuals (Harvey et al., 2021). At the time of writing, five variants have been characterized as VOC, each containing multiple specific spike mutations of interest: Alpha (Pango lineage B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) (“SARS-CoV-2, 2021).
1. Overview of five different VOCs
The first VOC to be reported was Alpha (B.1.1.7), which was first detected in September 2020 in the United Kingdom. Alpha’s major mutation of concern is N501Y. This mutation increases the affinity of the spike protein to human ACE 2 receptors, which leads to a potentially higher transmissibility and virulence (Rauseo and O’Halloran, 2021). In addition to the N501Y mutation, this variant contains multiple other mutations in the S gene, most importantly a A570D substitution (Leung et al., 2021). As of December 2020, a lineage known as B.1.1.7 + E484K has also been detected. This lineage contains a E484K mutation of specific concern because it can cause immune escape (Jangra et al., 2021). In September 2020, another SARS-CoV-2 variant, Beta (B.1.351), was detected in South Africa. Characteristic spike mutations for this variant are N501Y, E484K and K417N (Tang et al., 2021). Later that year, in December 2020, a third VOC, Gamma (P.1), was reported in Brazil. P.1 has the same N501Y and E484K mutations as Beta, but also an additional K417T mutation (Maggi et al., 2021). Both K417N and K417T mutations are associated with viral escape and resistance to neutralizing antibodies (Wibmer et al., 2021). In April 2021, another variant, Delta (B.1.617), arrived in Europe. This variant was first identified in India at the end of 2020 and consists of three sub-lineages, all of which carry the L452R substitution. Sub-lineages B.1.617.1 and B.1.617.3 also have an E484Q mutation, which can contribute to immune escape. Sub-lineage B.1.617.2 does not contain the E484Q mutation, but has an additional T478K mutation. Only sub-lineage B.1.617.2 is considered a VOC (“SARS-CoV-2, 2021), (“COVID-19 Weekly Epidemiological Update Global overview, 2021). In November 2021, a fifth VOC, Omicron (B.1.1.529), was detected in South Africa. Remarkably, this variant contains over 30 mutations in the S gene, which also occur in other variants and are linked to immune escape. Due to the high number of mutations in the spike protein, there is great concern that these mutations might undermine the effectiveness of the current vaccines and/or cause increased transmissibility (Callaway, 2021), (Maes, 2021).
2. The need for rapid detection of VOCs
Since new variants will continue to emerge, an ideal detection method should be able to quickly respond to newly emerging strains. For this purpose, we developed a Sanger sequencing-based test that provides sequence information in the receptor binding domain (RBD) region of the spike protein.
Primers were designed to ensure that most of the important mutations in the S gene are present in the amplicon that involves the RBD region. Our amplification allows the detection of mutations in the S gene between AA360 and AA588. Based on different combinations of mutations found in the sequence, all current VOCs can be differentiated. Table 1 lists these variants with their corresponding mutations within the region amplified using our method.
Table 1.
Overview of all the VOCs, with their characteristic mutations in the spike gene that can be detected in the 733 bp amplicon (“SARS-CoV-2 variants of concern as of 26 November 2021.”, 2021), (Gibson, 2011).
| WHO label | Pango Lineage | Spike mutations in the 733 bp amplicon | ||
|---|---|---|---|---|
| VOC | Alpha | B.1.1.7 | N501Y A570D |
|
| B.1.1.7 + E484K | E484K N501Y A570D |
|||
| Beta | B.1.351 | K417N E484K N501Y |
||
| Gamma | P.1 | K417T E484K N501Y |
||
| Delta | B.1.617.2 | L452R T478K |
||
| Omicron (BA.1) | B.1.1.529.1 | S371L S373P S375F K417N N440K G446S S477N T478K |
E484A Q493R G496S Q498R N501Y Y505H T547K |
|
As a validation, whole genome sequencing (WGS) by means of MinION sequencing (Oxford Nanopore Technologies, UK) was performed in parallel with the Sanger sequencing for 147 selected samples with different variants. The Sanger sequencing method was validated by comparing the results from both sequencing methods (Table S1). For all samples in this survey, the same outcome was obtained with the two methods. No discordance in mutations was observed, confirming the validity of this method as a trustworthy way to detect mutations in the S gene of SARS-CoV-2.
3. Detection of the first Omicron in Belgium
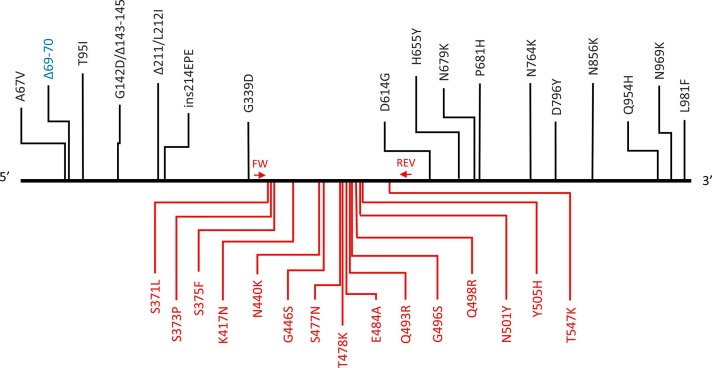
In case of the Omicron variant, an S gene target failure (SGTF) is typically observed in the commonly used TaqPath COVID RT-qPCR test (ThermoFisher) because of the AA69–70 deletion (Kidd et al., 2021). However, this 69–70 deletion also occurs in Alpha and in some Delta lineages, so sequencing should provide confirmation. On the 25th of November 2021, a sample with SGTF was noticed in the COVID-19 Federal Platform of UZ Leuven (Leuven, Belgium) and sent directly to the REGA Institute (KU Leuven, Leuven Belgium) for whole genome sequencing and Sanger sequencing. Both tests detected an Omicron variant (EPI_ISL_6794907), the first confirmed case in Belgium and Europe. Of the 32 S gene mutations present in the Omicron variant, 15 are present in the region covered by our Sanger sequencing method ( Fig. 1), hereby allowing reliable identification of this variant.
Fig. 1.
SARS-CoV-2 S-gene with indication of the amino acid mutations present in the Omicron variant and positions of primers used for Sanger sequencing. The mutation in blue is detected as S-gene target failure with the TaqPath COVID-19 RT-PCR test, mutations in red are detected with our Sanger sequencing method. Δ = deletion; ins=insertion; FW: forward primer (AA360); REV: reverse primer (AA588).
4. Methods
Viral RNA from positive nasopharyngeal swab samples was extracted from 200 µL of swab transporting medium using a Kingfisher Flex-96 (ThermoFisher Scientific, Europe). RT-PCR was performed to amplify a fragment of the S gene using the QIAGEN© One-step RT-PCR kit (QIAGEN ref. 210212). For amplification, a mastermix was made with 5 µL 5x OneStep RT-PCR Buffer, 1 µL OneStep RT-PCR enzyme mix, 1 µL dNTP mix (10 mM), 7 µL RNase free water, 3 µL forward primer (10 µM) and 3 µL reverse primer (10 µM). Five µL of viral RNA was added to the mix to reach a total of 25 µL. Primers were designed after performing an alignment of different variants in Mega v7.0: S-VOC-F (22618–22639) 5’-TTG GAA CAG GAA GAG AAT CAG C-3’ and S-VOC-R (23348–23324) 5’- TGA CAC CAC CAA AAG AAC ATG G − 3’. These primers generate an amplicon of 733 bp in the S gene coding for AA360 to AA588. Cycling conditions are 30 min at 50 °C, 15 min at 95 °C followed by 40 cycles of 30 s at 95 °C, 30 s at 55 °C and 1 min at 72 °C, and a final extension step of 10 min at 72 °C. Then 4 µL PCR product is purified with 2 µL H20 and 2 µL ExoSap-IT™ Product Cleanup Reagent (Thermo Fisher Scientific, ref. 78201.1.mL) by incubating at 37 °C for 15 min to degrade remaining primers and nucleotides, followed by an inactivation step at 80 °C for 15 min. Five µL of purified PCR product and 5 µL of S-VOC-F forward primer (5 µM) were mixed together in a 1.5 mL tube and sent to the Macrogen sequencing facility in Maastricht (The Netherlands) for Sanger sequencing on a 3730xl DNA Analyzer (ThermoFisher Scientific, Europe) (“Standard-Seq | Services | Macrogen Europe, 2022). The sequences were obtained within 24 h and manually corrected using Chromas 4.1. The resulting FASTA-files were analyzed with NextClade to identify signature mutations in the spike gene (Hodcroft, 2021). Nanopore sequencing was performed as described by Wawina-Bokalanga et al. (Wawina-bokalanga).
To demonstrate reproducibility, 3 samples were tested for Sanger sequencing and WGS in triplicate and also a cross-validation was made with 3 samples from other labs in Belgium. These results are shown in Table S1. The costs of Sangers sequencing and WGS were compared and listed in table S2.
5. Conclusion
For unambiguous typing of a SARS-CoV-2 positive sample, whole genome sequencing (WGS) is required. A major drawback of this approach is the high cost, especially when not batching samples. When performing batch sampling, the costs for whole genome sequencing can be significantly reduced. However, the cost remains higher than that of Sanger sequencing. Moreover, having to wait for sufficient samples to run complete batches, especially in smaller labs, can cause significant delays in reporting. Even without taking into account possible delays due to sample batching, WGS has a longer turnaround time (3–5 days on average) compared to Sanger sequencing (< 24 h with the method described here) due to a more laborious lab protocol and the need for a lengthy run on the sequencer in order to generate sufficient data. Additionally, there are extensive computational requirements, and a high level of expertise is needed. Sanger sequencing is an easy to use, fast and low-cost way to sequence SARS-CoV-2 samples and to distinguish different variants. A One-step RT-PCR reaction was developed that covers 288 amino acids in the RBD region of the spike gene, which encompasses most of the signature mutations hereby allowing variant identification. PCR amplicons can be sent to Macrogen (Europe) for fast Sanger sequencing. Because the Omicron variant has a lot of mutations in the S gene, the amplicon sequence allows unambiguous identification using NextClade. At the time of writing, the sub-lineage BA.2 (B.1.1.529.2) of Omicron had not been detected in Belgium yet, but since this sub-lineage contains extra key mutations (T376A/D405N/R408S) that are encompassed in the region that we sequence with our method, our assay should be able to unambiguously distinguish BA.2 from the original Omicron (now redesignated as B.1.1.529.1 or BA.1).
An alternative method for the detection of signature mutations of circulating VOCs is the use of a combination of multiplex qRT-PCR assays, targeting different specific mutations, which are carried out in parallel. This approach is suitable for implementation in a high-throughput setting, but the fact that this detection method is limited to a small number of specific mutations is a major drawback. Whenever new VOCs, carrying different mutations of interest, are detected, the test needs to be adapted, and new specific primer/probe sets need to be developed and validated. Our method, which is based on sequence determination of the region of interest of the spike RBD, does not have this drawback, making it a highly flexible tool for the detection of circulating variants.
CRediT authorship contribution statement
MB: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. AR: Visualization, Writing - review & editing. JS: Methodology, Validation. MVR: Conceptualization, Resources, Funding acquisition. PM: Methodology, Validation, Formal analysis, Investigation. BVM: Methodology, Validation, Formal analysis, Investigation, Writing – review & editing. EW: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – review & editing, Supervision
Additional information
This rapid communication is an original paper and has not been submitted elsewhere.
All authors have approved the final manuscript.
The corresponding author has read and agreed to the terms of the Eurosurveillance data protection notice.
The identification of SARS-CoV-2 variants is part of our function as Belgian National Reference Center for coronavirus diagnosis. An informed consent was therefore not necessary.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Reference Center for Respiratory pathogens from the RIZIV/INAMI (National Institute for health and Disability Insurance) and the HONOURs Horizon 2020 Marie Sklodowska-Curie Training Network (721367).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2022.114512.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- “COVID-19 Weekly Epidemiological Update Global overview.” Accessed May 18 2021. [Online]. Available: 〈https://reliefweb.int/sites/reliefweb.int/files/resources/20210511_Weekly_Epi_Update_39.pdf〉.
- “SARS-CoV-2 variants of concern as of 26 November 2021.” 〈https://www.ecdc.europa.eu/en/covid-19/variants-concern〉 (Accessed 1 December 2021.
- “Standard-Seq | Services | Macrogen Europe.” 〈https://www.macrogen-europe.com/services/sanger-sequencing/standard〉 (Accessed 10 February 2022).
- Callaway E. Heavily mutated coronavirus variant puts scientists on alert. Nature. 2021;600:21. doi: 10.1038/D41586-021-03552-W. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft , “Nextclade,” 2021. https://clades.nextstrain.org/ (Accessed 15 June 2021).
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Chernet R.L., Eaker L.Q., Ferreri E.D., Floda D.L., Gleason C.R., Kleiner G., Jurczyszak D., Matthews J.C., Mendez W.A., Mulder L., Russo K.T., Salimbangon A.T., Saksena M., Shin A.S., Sominsky L.A., Srivastava K. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microb. 2021:e283–e284. doi: 10.1016/s2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B., Mayhew M., Megram O., Ashford F., White T., Moles-Garcia E., Crawford L., Bosworth A., Atabani S.F., Plant T., McNally A. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J. Infect. Dis. 2021;223(10):1666–1670. doi: 10.1093/INFDIS/JIAB082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance. 2021;26(1) doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes, P. , Genomic surveillance of SARS-CoV-2 in Belgium,” Accessed 29 November 2021. [Online]. Available: 〈https://www.uzleuven.be/nl/laboratoriumgeneeskunde/genomic-surveillance-sars-cov-2-belgium〉.
- Maggi F., Novazzi F., Genoni A., Baj A., Spezia P.G., Focosi D., Zago C., Colombo A., Cassani G., Pasciuta R., Tamborini A., Rossi A., Prestia M., Capuano R., Azzi L., Donadini A., Catanoso G., Grossi P.A., Maffioli L., Bonelli G. Imported SARS-COV-2 Variant P.1 Detected in Traveler Returning from Brazil to Italy. Emerg. Infect. Dis. 2021;27(4):1249–1251. doi: 10.3201/eid2704.210183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauseo A.M., O’Halloran J.A. What are the clinical implications of the SARS-CoV-2 variants. JACC Basic Transl. Sci. 2021;6:305–308. doi: 10.1016/j.jacbts.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.007. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawina-bokalanga, T. , Genetic diversity and evolution of SARS- CoV-2 in Belgium during the first wave outbreak.”.
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J.N., Moore P.L. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material