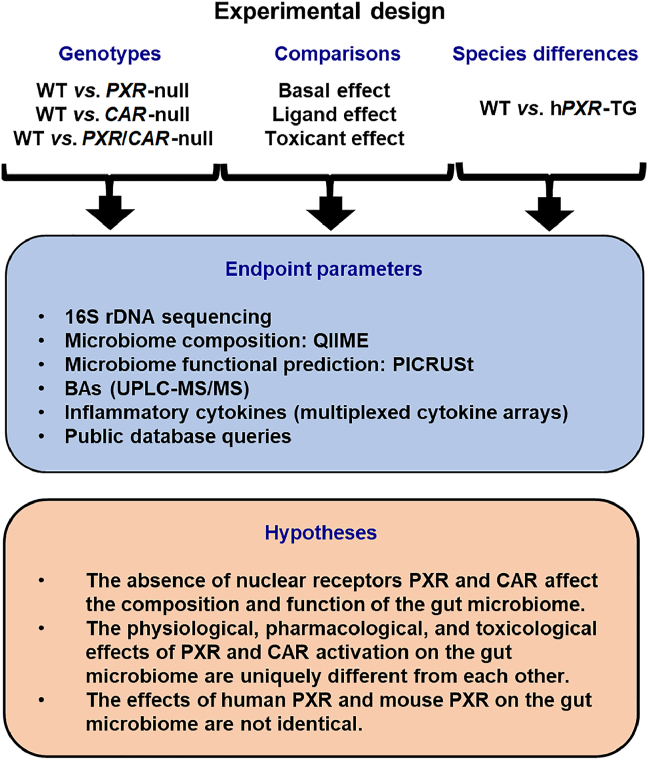
Figure 1.
Schematic of the experimental design of this study. [Study 1] To determine the necessity of the host PXR and CAR genes on modulating the compositions and functions of the gut microbiome under physiological conditions, fecal samples were collected from WT, PXR-null, CAR-null, and PXR-CAR-null (all in C57BL/6 background) male and female mice of adolescent and adult ages (n = 5 of each group). [Study 2] To compare the physiological roles of PXR and CAR with ligand-mediated activation and toxicant-mediated activation of these nuclear receptors, data from the present study were compared with publicly inquired databases of the same genetic background (C57BL/6), sexes, and age. For ligand effect, 16S rDNA sequencing data were retrieved from mice that were orally exposed to the prototypical PXR ligand PCN or the prototypical CAR ligand TCPOBOP30. For toxicant effect, 16S rDNA sequencing data were retrieved from mice that were orally exposed to PBDEs9,13,37 or PCBs31,40 [Study 3] To compare the role of mouse and human PXR genes on the composition and function of the gut microbiome, fecal samples were collected from WT and hPXR-TG (both in FVB background) male and female mice of adolescent and adult ages (n = 5 of each group). Due to poor breeding capacity of hCAR-TG mice in C57BL/6 background and the lack of access of hCAR-TG mice in the FVB background, the comparison of mouse and human CAR genes on gut microbiome was not determined in this study. Fecal samples were collected after 24 h, and 16S rDNA gene sequencing was conducted by amplifying the hypervariable V4 region. Analysis of FASTQ files was conducted using various python scripts in Quantitative Insights Into Microbial Ecology (QIIME)41, including de-multiplexing, quality filtering, operational taxonomy unit (OTU) picking, as well as alpha- and beta-diversity determinations. Metagenome functional content was predicted using Phylogenetic Investigation of Communities by Reconstruction of Observed States (PICRUSt)42. BAs were quantified using liquid chromatography–tandem mass spectrometry (LC–MS/MS). Cytokines were quantified using the Mouse Cytokine Array Pro-inflammatory Focused 10-Plex (MDF10; Eve Technologies Corp., Calgary, Alberta, Canada). For Study 1, we hypothesize that the absence of PXR and CAR affect the composition and function of the gut microbiome. For Study 2, we hypothesize that the physiological, pharmacological, and toxicological effects of PXR and CAR activation on gut microbiome are uniquely different from each other. For Study 3, we hypothesize that the effects of human PXR and mouse PXR on the gut microbiome are not identical.