Abstract
A resurging interest in targeted covalent inhibitors (TCIs) focus on compounds capable of irreversibly reacting with nucleophilic amino acids in a druggable target. p97 is an emerging protein target for cancer therapy, viral infections and neurodegenerative diseases. Extensive efforts were devoted to the development of p97 inhibitors. The most promising inhibitor of p97 was in phase 1 clinical trials, but failed due to the off-target-induced toxicity, suggesting the selective inhibitors of p97 are highly needed. We report herein a new type of TCIs (i.e., FL-18) that showed proteome-wide selectivity towards p97. Equipped with a Michael acceptor and a basic imidazole, FL-18 showed potent inhibition towards U87MG tumor cells, and in proteome-wide profiling, selectively modified endogenous p97 as confirmed by in situ fluorescence scanning, label-free quantitative proteomics and functional validations. FL-18 selectively modified cysteine residues located within the D2 ATP site of p97. This covalent labeling of cysteine residue in p97 was verified by LC‒MS/MS-based site-mapping and site-directed mutagenesis. Further structure–activity relationship (SAR) studies with FL-18 analogs were established. Collectively, FL-18 is the first known small-molecule TCI capable of covalent engagement of p97 with proteome-wide selectivity, thus providing a promising scaffold for cancer therapy.
Key words: p97, Activity-based protein profiling, Targeted covalent inhibitor, Chemical proteomics
Graphical abstract
Despite many inhibitors of p97 have been reported recently, its target covalent inhibitors (TCIs) are still not available. We report herein a p97-targeting TCI (FL-18) showing unprecedented proteome-wide selectivity.
1. Introduction
Many cellular proteins, including cysteine proteases1 and kinases2, participate in essential biological functions by using nucleophilic active-site residues such as cysteine and lysine. Drug discovery scientists over the years have been able to take advantage of such nucleophiles by developing highly selective targeted covalent inhibitors (TCIs) that incorporate corresponding electrophiles via rational design3. Despite initial concerns of promiscuity and off-targeting, many of these TCIs have shown good on-target selectivity and some are already being used in the clinic4. As such, there has been a resurgence of interests in the development of TCIs with novel electrophiles5,6. The α,β-unsaturated Michael acceptors are amongst the most commonly used electrophiles employed in TCIs, capable of selectively targeting cysteine residues in a protein target7.
VCP/p97 AAA ATPase (henceforth named p97) is a homohexameric enzyme in mammalian cells that works as a molecular engine to extract ubiquitinated proteins from protein complexes and organelles, and facilitates their subsequent degradation by ubiquitin proteasome system (UPS)8,9. It consists of three domains, the N-terminal domain, D1 and D2 domains10,11. Recent studies have shown that inhibition of p97 provides excellent therapeutic effect in hematological and solid tumor models12,13. A number of promising p97 inhibitors have been reported, capable of inhibiting the enzyme either reversibly12,14, 15, 16, 17, 18 or irreversibly19, 20, 21. The most promising inhibitor of p97 was in phase 1 clinical trials, but failed due to the off-target-induced toxicity22, suggesting the selective inhibitors of VCP/p97 are highly needed. Herein, we report the first-ever covalent inhibitor (i.e., FL-18) that shows proteome-wide selectivity towards endogenous p97. Notably, FL-18 covalently labeled cysteine residues in the catalytic domain of p97 and showed potent enzymatic inhibition. The discovery process of TCI FL-18 and its unique mode of action with its protein target will provide a new design paradigm to develop other TCIs in future.
2. Materials and methods
The detailed description of materials and methods except the following two methods is provided in the Supporting Information.
2.1. Validation of the labeling between p97 and J-1 probe
U87MG cells were grown to 70% confluency in 10-cm dishes and incubated with 1 μmol/L probe J-1 at 37 °C with 5% CO2 for 1 h. Cells were harvested, lysed in 0.1% NP-40/PBS using a probe sonicator, and centrifuged at 15,000 rpm for 30 min to remove cell debris. The concentrations of the whole proteome were determined by BCA protein assay and normalized to 2 mg/mL. Click chemistry was performed on each sample by using final concentrations of 5 μmol/L TAMRA-biotin-azide and other reagents (Tris (2-carboxyethyl) phosphine (TCEP, Sigma–Aldrich), Tris [(1-benzyl-1H-1,2,3-triazol-4-yl) methyl] amine (TBTA, Sigma–Aldrich) and CuSO4) in a final volume of 1 mL for 1 h. After click chemistry, the proteins were precipitated at 6500 ×g for 5 min. Then the cold methanol was used to wash the precipitated sample and the lysate was fractionated by centrifugation at 6500 ×g for 5 min at 4 °C. The precipitated protein pellets were resuspended and dissolved in 1.2% SDS/PBS (1 mL final volume). The proteomes were boiled at 90 °C for 5 min, and after centrifugation at 1400 ×g for 1 min at room temperature, the supernatant was diluted to 0.2% SDS/PBS. Streptavidin beads were washed with PBS for 3 times and the above supernatant was enriched with beads for 3 h at 29 °C. The beads were washed with PBS for 3 times, and washed with H2O for 3 times. The beads were boiled in 50 μL of 1× gel-loading buffer at 95 °C for 20 min, which resulted in successful elution of the bead-bound proteins.
In pull-down experiments, the eluted samples were separated on a 10% SDS-PAGE gel and immunoblotted with anti-p97 antibody (ab111740).
In gel LC‒MS/MS experiments, the eluted samples were separated on a 10% SDS-PAGE gel, followed by in-gel trypsin digestion. The resulting peptides were extracted and desalted for downstream LC‒MS/MS analysis.
2.2. Mapping inhibitor-reactive amino acid by LC‒MS/MS
Recombinant p97 protein (50 μL, 10 μmol/L) in PBS buffer (pH 7.4) was incubated with the corresponding inhibitor (FL-18, WK3429 or WK3429, 1 μL, 10 μmol/L) for 1 h at room temperature. To the resulting mixture was then added 150 μL of 8 mol/L urea and 100 mmol/L DTT, and the mixture was incubated for 1 h at 35 °C. Then the sample was transferred to a 30-kDa cut-off filter (Microcon-30 kDa Centrifugal Filter Unit with Ultracel-30 membrane, Millipore). After centrifugation at 14,000 ×g for 20 min, the sample was washed with 150 μL of 50 mmol/L NH4HCO3 at 14,000 ×g for 20 min. Then 20 mmol/L IAA (150 μL) was added to the filter, followed by further incubation for 1 h in dark. After centrifugation at 14,000 ×g for 20 min, the sample was washed with 150 μL of 50 mmol/L NH4HCO3 for three times. Next, 150 μL of 10 mmol/L NH4HCO3 and trypsin (trypsin:protein = 1:100, w/w) to the sample, followed by further incubation at 37 °C for 17 h. The resulting peptides were extracted and desalted for downstream LC‒MS/MS analysis.
3. Results and discussion
3.1. FL-18 and its activity-based probe showed potent inhibition towards U87MG glioma cancer cells
Our study started from a random high-throughput screening campaign that led to the serendipitous discovery of FL-18 (Fig. 1A, see Supporting Information Scheme S1 for synthesis) with low nanomolar inhibition towards U87MG glioma cancer cells (IC50 = 31 nmol/L; Fig. 1B). We were excited by this discovery as FL-18 may have potential therapeutic benefit to human gliomas (a lethal neoplasm with a poor prognosis)23. However, the exact protein targets of FL-18 was not clear. We thereby synthesized a terminal alkyne-containing clickable molecular probe (J-1; Fig. 1C), which could be employed to capture the probe-modified proteins from live mammalian cells via copper (I)-catalyzed alkyne–azide cycloaddition (CuAAC)―a process that is now commonly referred to as activity-based protein profiling (ABPP)24, 25, 26.
Figure 1.
FL-18 effectively kills U87MG glioma cells. (A) The chemical structures of FL-18. (B) Viability of U87MG cells upon incubation with FL-18 for 48 h. (C) The synthetic route of J-1 probe. a) trimethyl orthoformate, PTSA, MeOH, rt, 5 h; b) propynol ethoxylate, NaOH, 70 °C, 8 h; c) HCl, acetone, rt, 2 h; d) ethylene diamine, t-BuOH, rt, 1 h, then K2CO3, I2, 70 °C, 3 h; e) iodobenzene diacetate, K2CO3, DMSO, rt; f) Zn, NH4Cl, MeOH, H2O, reflux, 1 h; g) 3-phenylpropargyl chloride, pyridine, THF, rt, 3 h. (D) Viability of U87MG cells upon incubation with J-1 probe for 48 h.
The probe J-1 was synthesized in 7 steps (Scheme S1), and unequivocally verified by HRMS and NMR. Similar to FL-18, probe J-1 possessed potent inhibition toward U87MG cells (IC50 = 27 nmol/L, Fig. 1D), indicating the introduction of an alkyne handle had negligible effect on the biological activity of FL-18. Thus, J-1 could serve as a molecular probe to reveal potential endogenous protein targets of FL-18.
3.2. Protein target identification of FL-18 via activity-based protein profiling
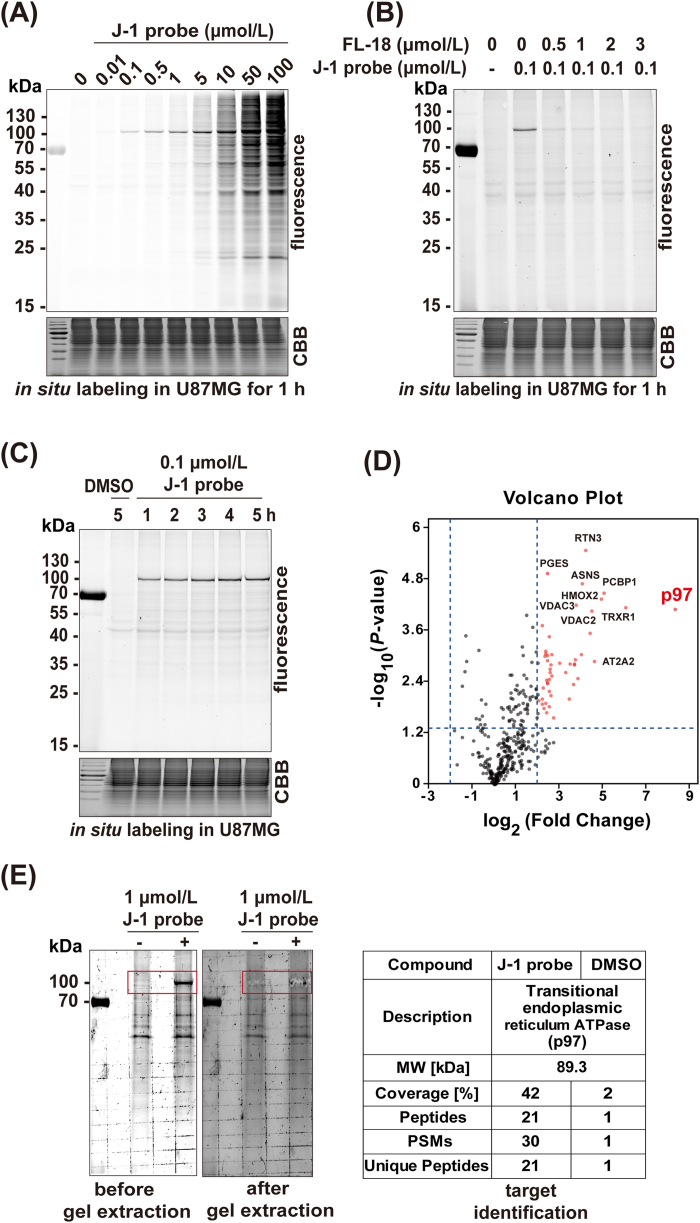
We next employed three chemical proteomic approaches to examine potential protein targets of FL-18 in live mammalian cells with J-1 (Fig. 2). First, we treated live U87MG cells with J-1 for 1 h, followed by lysis and click reaction with rhodamine-azide (Fig. 2, top approach). Upon SDS-PAGE and in-gel fluorescence scanning of the labeled proteomes, a highly selectively labeled band at ∼100 kDa emerged when the concentration of J-1 was <1 μmol/L (Fig. 3A); at higher probe concentrations (>5 μmol/L), other labeled bands started to appear, indicating off-targeting. Given the fact that this 100-kDa probe-labeled band could be successfully blocked by preincubation of FL-18 with live cells (Fig. 3B), and both FL-18 and J-1 possessed submicromolar U87MG-killing activities, our results thus indicate the 100-kDa band likely represented the genuine cellular target of FL-18. In addition, J-1-induced fluorescence labeling was shown to follow a time-dependent manner with saturated labeling at 4 h (Fig. 3C).
Figure 2.
The three approaches employed to identify the true cellular protein target of FL-18.
Figure 3.
FL-18 selectively targeted p97 in U87MG cancer cells. (A) In situ concentration-dependent ABPP labeling of U87MG cells with J-1 probe. (B) Competitive labeling of J-1 probe with FL-18 in U87MG cells. (C) Time-dependent labeling of J-1 in U87MG cells. (D) Volcano plot of gel-free quantitative LC‒MS experiments of proteomes labeled with J-1 (1 μmol/L) or DMSO (negative control). (E) In situ labeling of U87MG cells with J-1, followed by CuAAC with a trifunctional reporter, pull-down (PD) and in-gel digestion of J-1-enriched 100-kDa labeled band, and finally LC‒MS/MS analysis.
Subsequently, two additional chemical proteomic approaches were employed to accurately elucidate the identity of the 100-kDa labeled protein. First, we treated the J-1-labled proteome with a biotin-azide reporter via CuAAC, followed by affinity enrichment (i.e., pull-down) and label-free quantification (Fig. 2, middle approach). By comparing the fold changes between the J-1-captured and vehicle-captured proteomes, we found that, amongst all captured proteins identified, the p97 protein showed the largest log2 (fold change) (8.3) (Fig. 3D). With a molecular weight of 89.3 kDa, a probe-labeled p97 would be expected to show a similar migration pattern as the 100-kDa labeled band from our earlier SDS-PAGE/in-gel fluorescence scanning. To further confirm our label-free quantification results and assign the 100-kDa band unequivocally to p97, we next performed another commonly used proteomic method. By treating the J-1-labeled proteome via CuAAC with a trifunctional reporter containing both biotin and rhodamine, followed by pull-down with avidin beads (Fig. 2, bottom approach), the enriched proteome was separated by SDS-PAGE. We then cut off the 100-kDa labeled band and subsequently performed in-gel digestion and LC‒MS/MS analysis. As shown in Fig. 3E (right table), 21 unique peptides covering 42% of the entire p97 protein sequence were detected in the 100-kDa J-1-labeled band, whereas only one peptide was detected in the control experiment. Thus, this result was consistent with that of the label-free quantification approach, confirming that endogenous p97 was indeed the genuine on-target of FL-18/J-1 under biologically relevant conditions.
3.3. The functional validation of the interaction between p97 and FL-18
We next performed multiple biochemical assays to functionally validate the proteomic results (Fig. 4). First, the pull-down analysis was done by using anti-p97 antibody, indicated that J-1 could efficiently enrich the p97 protein from the whole proteome (Fig. 4A); this enrichment was abolished by preincubation of the whole proteome in the presence of FL-18, consistent with the above-mentioned in-gel fluorescence scanning. Furthermore, knock-down of p97 with siRNA greatly decreased the fluorescence intensity of the J-1-labeled band (Fig. 4B and Supporting Information Fig. S1). In addition, the recombinant p97 could be efficiently labeled by J-1 in a concentration-dependent manner (Fig. 4C). The direct biological function of p97 is to convert ATP to ADP27. We next checked the enzymatic activity of p97 in the presence of FL-18, by measuring the concentration of byproduct (phosphate) during the p97-catalyzed ATP-to-ADP conversion (Supporting Information Fig. S2A). As shown in Fig. 4D, FL-18 could potently inhibit the activity of p97 with an IC50 value of 59.3 nM. The measured value of Kinact/Ki was 0.0123 L/nmol·min and 0.0098 L/nmol·min for FL-18 and J-1, respectively (Supporting Information Fig. S3). In addition, FL-18 showed better inhibition potency than NMS-873, a well-known p97 inhibitor (Fig. S2B)19. The biological effect of p97 is to promote the degradation of ubiquitinated proteins10. And we found that the knock-down of p97 with siRNA significantly induced the buildup of ubiquitinated proteins (Fig. 4E). Notably, cells treated with J-1 had the same effect as those treated with p97 siRNA or a proteasome inhibitor (Cfz; Fig. 4F). Next, we checked if J-1 could label other ATPases. We selected and purified N-ethylmaleimide sensitive factor (NSF), a close homologue of p97. As shown in Supporting Information Fig. S4, J-1 could not label NSF, or FL-18 did not affect the activity of NSF (Supporting Information Fig. S5). Together, these functional validation experiments undoubtedly supported the specific covalent interaction between FL-18 and endogenous p97 from tumor cells.
Figure 4.
FL-18 inhibited the p97 activity and induced aggregation of ubiquitinated proteins. (A) Competitive labeling of p97 by J-1 probe with FL-18. The labeled proteomes were “clicked” with biotin reporter (‘input’), pulled down by avidin beads and eluted by boiling the beads (‘output’). The input and output samples were analyzed by anti-p97 immunoblotting. (B) Labeling of endogenous p97 by J-1 in U87MG cells, with or without small-interfering RNA (siRNA p97). CBB is shown as a loading control. (C) Concentration-dependent labeling of recombinant p97 by J-1. (D) FL-18 efficiently inhibited the enzymatic activity of recombinant p97. (E) Endogenous p97 knockdown induced aggregation of ubiquitinated proteins. (F) J-1 efficiently induced aggregation of ubiquitinated proteins. Cfz, carfilzomib (a proteasome inhibitor). CBB, Coomassie brilliant blue; UB, ubiquitinated proteins.
3.4. FL-18 covalently modified C522 residue in p97
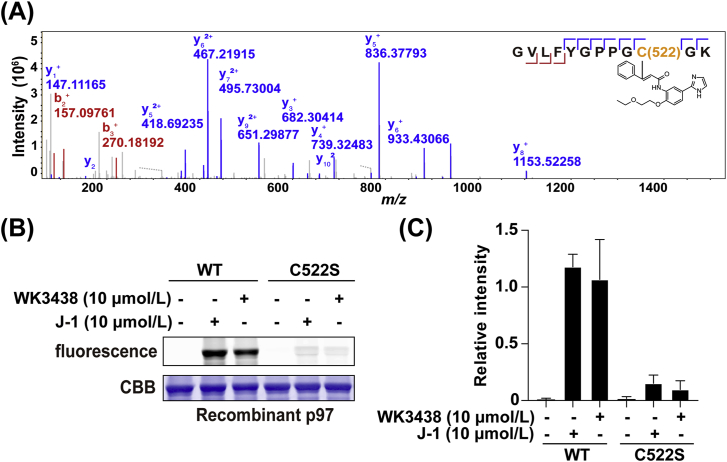
We subsequently analyzed the direct binding site of FL-18 in p97. To do so, we carried out LC‒MS/MS analysis of peptides generated by tryptic digestion of FL-18-treated recombinant p97. The site-mapping results indicated that FL-18 could label Cys522 of p97 (Fig. 5A), which was consistent with molecular docking that is suggestive of a close distance between Cys522 and the Michael acceptor in FL-18 (Supporting Information Fig. S6). To further confirm the targeting behavior of this novel probe, we next carried out the labeling experiment using both wild-type and p97 mutants. We first prepared recombinant C522S mutant of p97 by site-directed mutagenesis and subsequently expressed/purified the resulting proteins, followed by J-1 labeling. As shown in Fig. 5B and C, the labeling intensity of C522S mutant by J-1 decreased significantly when compared to that of the wild-type p97 under identical labeling conditions, only a faint band could be observed (Supporting Information Fig. S7). Furthermore, similar labeling profiles, as shown in Figs. 5B, C and S4, were observed for these p97 mutants with WK3438, an analog of J-1. Together, these results clearly demonstrated that FL-18 could label the C522 residue in p97.
Figure 5.
Potential sites of p97 covalently engaged by FL-18. (A) Representative MS/MS site-mapping data showing the identification of Cys522 as the main reacting site of FL-18 in recombinant p97. (B) The fluorescence labeling of wide-type and mutant p97 by J-1 and WK3438. (C) The relative band intensity of labelled p97 by J-1 and WK3438 (n = 2 independent experiments).
3.5. Target-directed structure–activity-relationship studies
Finally, we carried out structure–activity relationship (SAR) studies by generating FL-18 analogs with structural variations at the flanking phenyl and imidazole rings. For the part of phenyl ring, we also synthesized compound WK3429, in addition to above-mentioned WK3438 (Scheme S1, Fig. 6A). WK3438 contained an electron-donating methoxyl group while WK3429 contained an electron-withdrawing trifluoromethyl group. The in-gel fluorescence scanning showed that WK3429 labeled more proteins than both WK3438 and J-1 (Fig. 6B). The site-mapping analysis of recombinant p97 labeled by WK3438 and WK3429 indicated that both compounds modified Cys522 in p97 (Supporting Information Figs. S8 and S9). Together, these results indicated that the additional electron-withdrawing group could render the α,β-unsaturated amide more reactive, while the electron-donating group had led to attenuation of reactivity. To our surprise, we observed that WK3429, amongst the three tested compounds (Fig. 6C), exhibited the worst growth inhibition of U87MG cells, indicating that excessive reactivity or non-specific labeling could have compromised the p97-targeting property of FL-18 analogs. We also synthesized two analogs of FL-18, WK4119 and WK4121, which contained a thiazole group and thiophene group, respectively (Supporting Information Schemes S2 and S3, Fig. 6D). As shown in Fig. 6E, both compounds had comparatively weaker inhibitory properties on p97 when compared to FL-18. In these two compounds, the number of nitrogen atom in the original imidazole ring of FL-18 was intentionally removed sequentially (e.g., N = 1 in WK4119; N = 0 in WK4121). Interestingly, we observed a corresponding decrease in the inhibition of p97 activity implying the basicity of imidazole was prefered for the ligand to bind and react with p97.
Figure 6.
Effect of structural variations on the bioactivity of FL-18. (A) Chemical structures of WK3438 and WK3429. (B) In situ concentration-dependent labeling of J-1, WK3438 and WK3429 in U87MG cells, respectively. (C) Cell viability of U87MG cells upon incubation with FL-18, WK3438 or WK3429 for 72 h. (D) Chemical structures of WK4119 and WK4121. (E) Inhibition of p97 enzymatic activity by FL-18, WK4119 or WK4121.
4. Conclusions
In summary, we have employed a plethora of chemical biology experiments to confirm that FL-18 is a novel TCI that could selectively label p97 covalently in live tumor cells, leading to subsequent potent inhibition of cell growth of gliomas. Notably, FL-18 and its analogs could label the key cysteine reside within the same active site of p97. FL-18 is thereby a promising scaffold for cancer therapy. In addition, our preliminary structure–activity relationship studies with analogs of FL-18 may shed light for further structural optimizations of this unique class of p97-targeting inhibitors.
Acknowledgments
We thank the funding support from Institute of Materia Medica, Peking Union Medical College, CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-4-005, China), The Natural Science Foundation of China (No. 22177136), and the Synthetic Biology Research & Development Programme (SBP) of National Research Foundation (SBP-P4 and SBP-P8) of Singapore.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.09.003.
Contributor Information
Zhiqiang Feng, Email: fengzhiq@imm.ac.cn.
Chong-Jing Zhang, Email: zhangchongjing@imm.ac.cn.
Author contributions
Zhiqiang Feng and Chong-Jing Zhang conceived and designed the study. Zi Ye and Lianguo Chen performed target identification and validation. Ke Wang and Xiaofeng Jin synthesized the compounds. Hao Chen and Guanghui Tang conducted molecule docking. Shao Q Yao provided useful suggestions and revised the manuscript. Chong-Jing Zhang, Zhiqiang Feng, Zi Ye, Ke Wang, Lianguo Chen wrote the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Olson O.C., Joyce J.A. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15:712–729. doi: 10.1038/nrc4027. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson F.M., Gray N.S. Kinase inhibitors: the road ahead. Nat Rev Drug Discov. 2018;17:353. doi: 10.1038/nrd.2018.21. [DOI] [PubMed] [Google Scholar]
- 3.Baillie T.A. Targeted covalent inhibitors for drug design. Angew Chem Int Ed. 2016;55:13408–13421. doi: 10.1002/anie.201601091. [DOI] [PubMed] [Google Scholar]
- 4.De Cesco S., Kurian J., Dufresne C., Mittermaier A.K., Moitessier N. Covalent inhibitors design and discovery. Eur J Med Chem. 2017;138:96–114. doi: 10.1016/j.ejmech.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Martín-Gago P., Olsen C.A. Arylfluorosulfate-based electrophiles for covalent protein labeling: a enw addition to the arsenal. Angew Chem Int Ed. 2019;58:957–966. doi: 10.1002/anie.201806037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray S., Murkin A.S. New electrophiles and strategies for mechanism-based and targeted covalent inhibitor design. Biochemistry. 2019;58:5234–5244. doi: 10.1021/acs.biochem.9b00293. [DOI] [PubMed] [Google Scholar]
- 7.Sutanto F., Konstantinidou M., Dömling A. Covalent inhibitors: a rational approach to drug discovery. RSC Med Chem. 2020;11:876–884. doi: 10.1039/d0md00154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura T., Wilkinson A.J. AAA+ superfamily ATPases: common structure-diverse function. Gene Cell. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 9.Meyer H., Bug M., Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S.L., Yan Y., et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan M., Zheng Q., Yu Y., Ai H., Xie Y., Zeng X., et al. Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative. Nat Commun. 2021;12:121. doi: 10.1038/s41467-020-20359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H.J., Wang J., Yao B., Wong S., Djakovic S., Kumar B., et al. Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083) J Med Chem. 2015;58:9480–9497. doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]
- 13.Huryn D.M., Kornfilt D.J.P., Wipf P. p97: an emerging target for cancer, neurodegenerative diseases, and viral infections. J Med Chem. 2020;63:1892–1907. doi: 10.1021/acs.jmedchem.9b01318. [DOI] [PubMed] [Google Scholar]
- 14.Chou T.F., Brown S.J., Minond D., Nordin B.E., Li K., Jones A.C., et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pöhler R., Krahn J.H., van den Boom J., Dobrynin G., Kaschani F., Eggenweiler H.M., et al. A non-competitive inhibitor of VCP/p97 and VPS4 reveals conserved allosteric circuits in Type I and II AAA ATPases. Angew Chem Int Ed. 2018;57:1576–1580. doi: 10.1002/anie.201711429. [DOI] [PubMed] [Google Scholar]
- 16.Figuerola-Conchas A., Saarbach J., Daguer J.P., Cieren A., Barluenga S., Winssinger N., et al. Small-molecule modulators of the ATPase VCP/p97 affect specific p97 cellular functions. ACS Chem Biol. 2020;15:243–253. doi: 10.1021/acschembio.9b00832. [DOI] [PubMed] [Google Scholar]
- 17.Wang F., Li S., Gan T., Stott G.M., Flint A., Chou T.F. Allosteric p97 inhibitors can overcome resistance to ATP-competitive p97 inhibitors for potential anticancer therapy. Chem Med Chem. 2020;15:685–694. doi: 10.1002/cmdc.201900722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan M., Yu Y., Ai H., Zheng Q., Xie Y., Liu L., et al. Mechanistic insight into substrate processing and allosteric inhibition of human p97. Nat Struct Mol Biol. 2021;28:614–625. doi: 10.1038/s41594-021-00617-2. [DOI] [PubMed] [Google Scholar]
- 19.Magnaghi P., D'Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 20.Wijeratne E.M., Gunaherath G.M., Chapla V.M., Tillotson J., de la Cruz F., Kang M., et al. Oxaspirol B with p97 inhibitory activity and other oxaspirols from lecythophora sp. FL1375 and FL1031, endolichenic fungi inhabiting parmotrema tinctorum and cladonia evansii. J Nat Prod. 2016;79:340–352. doi: 10.1021/acs.jnatprod.5b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding R., Zhang T., Wilson D.J., Xie J., Williams J., Xu Y., et al. Discovery of irreversible p97 inhibitors. J Med Chem. 2019;62:2814–2829. doi: 10.1021/acs.jmedchem.9b00144. [DOI] [PubMed] [Google Scholar]
- 22.Leinonen H., Cheng C., Pitkänen M., Sander C.L., Zhang J., Saeid S., et al. A p97/valosin-containing protein inhibitor drug CB-5083 has a potent but reversible off-target effect on phosphodiesterase-6. J Pharmacol Exp. 2021;378:31–41. doi: 10.1124/jpet.120.000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J., Lv X., Lu C., Ye X., Chen X., Fu J., et al. Prognostic factors of patients with gliomas―an analysis on 335 patients with glioblastoma and other forms of gliomas. BMC Cancer. 2020;20:35. doi: 10.1186/s12885-019-6511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones L.H. Reactive chemical probes: beyond the kinase cysteinome. Angew Chem Int Ed. 2018;57:9220–9223. doi: 10.1002/anie.201802693. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Patricelli M.P., Cravatt B.F. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu T., Li Y., Lu W., Spitters T., Fang X., Wang J., et al. Discovery of a subtype-selective, covalent inhibitor against palmitoylation pocket of TEAD3. Acta Pharm Sin B. 2021;11:3206–3219. doi: 10.1016/j.apsb.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T.F., Bulfer S.L., Weihl C.C., Li K., Lis L.G., Walters M.A., et al. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J Mol Biol. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.