Figure 7: EBF1 derepression confers GSI-resistance in T-ALL.
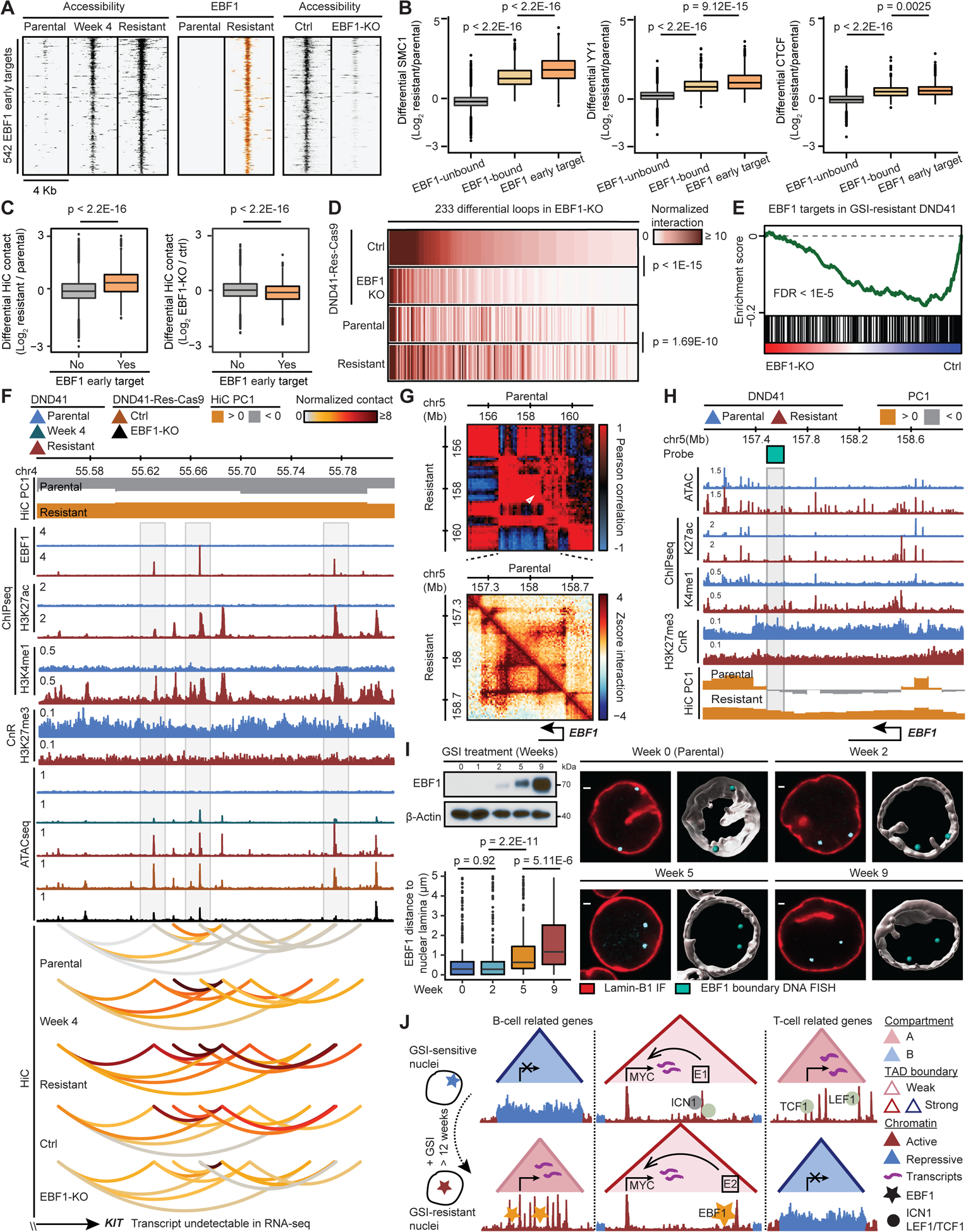
(A) Gradual gain and loss of opening during resistance acquisition and EBF1 deletion (EBF1-KO), respectively, at EBF1 early target elements.
(B) Architectural proteins increase the most at the EBF1 early target elements. EBF1-unbound: EBF1-unbound accessible elements in resistant DND41; EBF1-bound: resistant EBF1-bound elements without change of accessibility in EBF1-KO; EBF1 early target elements per (A) (Wilcoxon test).
(C) Differential HiC signal during resistance development (left) and post EBF1 deletion (right) per loop connected to EBF1 early target element (Wilcoxon test).
(D) HiC signal at EBF1-dependent loops connected to EBF1 early target elements (paired t-test).
(E) Repression of GSI-resistant EBF1 targets (Figure S4K) upon EBF1 deletion in DND41-Res-Cas9.
(F) KIT-connected EBF1-bound elements with EBF1-dependent opening and looping (gray boxes) showing chromatin restructuring can precede expression (STAR Methods).
(G) EBF1 locus Pearson correlation (top) and zoomed-in HiC contact frequency (bottom) matrices.
(H) EBF1 acquired active chromatin state and compartmentalization in resistant cells. Gray box: Oligopaint probe at EBF1-containing TAD boundary.
(I) Gradual EBF1 activation (left, top) and increase of distance between EBF1 and nuclear envelope (left, bottom) during resistance acquisition. Mean (+/− SD) distance at week 0 to 9: 0.74 (+/− 1.19), 0.72 (+/− 1.20), 1.18 (+/− 1.35) and 1.67 (+/− 1.44) μm. Right: representative and Imaris-modeled cells.
(J) Dissociation of B-lineage determining transcription factor EBF1 from nuclear lamina during GSI-resistance development instructs widespread and concerted chromatin refolding that regulates key genes including MYC to circumvent Notch-addiction in T-ALL.
