Figure 6.
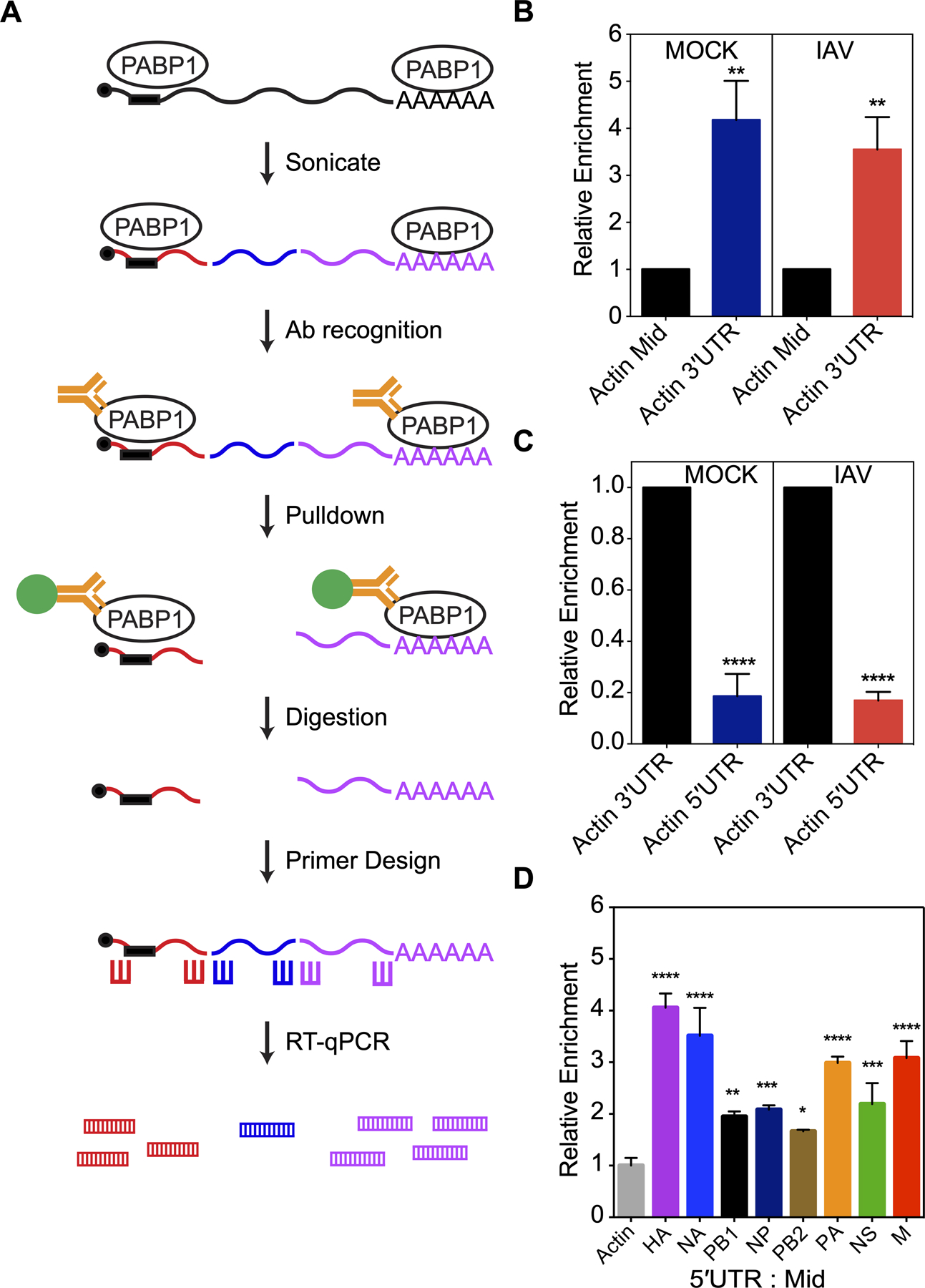
PABP1 pulldown from IAV-infected cells enriches for viral 5ʹUTR RNAs. (A) The workflow of the immunoprecipitation experiment from live IAV-infected cells. Briefly, lysate containing PABP1 crosslinked to RNA are sheared into fragments by the sonication step, followed by recognition of PABP1 by specific antibody and pulldown of the PABP1•RNA complex. Proteins are digested and total RNA fragments are purified. Primers for reverse transcription and RT-qPCR were designed to amplify the 5ʹ-, 3ʹ- and middle section of the mRNAs of interest. RT-qPCR was performed to determine the enrichment for the 5ʹ-, 3ʹ- and middle section of selected mRNAs. (B) Results of RT-qPCR comparing the enrichment of actin mRNA fragments from the 3ʹ-end (Actin 3ʹUTR) relative to the middle fragments (Actin Mid) set to 1. MOCK refers to cells that were mock infected while IAV refers to cells infected with A/Puerto Rico/8/34 (H1N1) IAV strain. (C) Results of RT-qPCR comparing the enrichment of actin mRNA fragments from the 5ʹ-end (Actin 5ʹUTR) relative to fragments from the 3ʹ-end (Actin 3ʹUTR) set to 1. MOCK refers to cells that were mock infected while IAV refers to cells infected with A/Puerto Rico/8/34 (H1N1) IAV strain. (D) Results of RT-qPCR comparing the enrichment of RNA fragments from the 5ʹ-end relative to fragments from the middle of IAV mRNAs. The data was normalized relative to the actin mRNA fragments from the 5ʹ-end over middle set to 1 and the error was propagated for all conditions. The cells were infected with A/Puerto Rico/8/34 (H1N1). The standard deviations from three biological replicates are shown. P < 0.05 is denoted with one asterisk, P < 0.01 is denoted with two asterisks, P < 0.001 is denoted with three asterisks, and P < 0.0001 is denoted with four asterisks.
