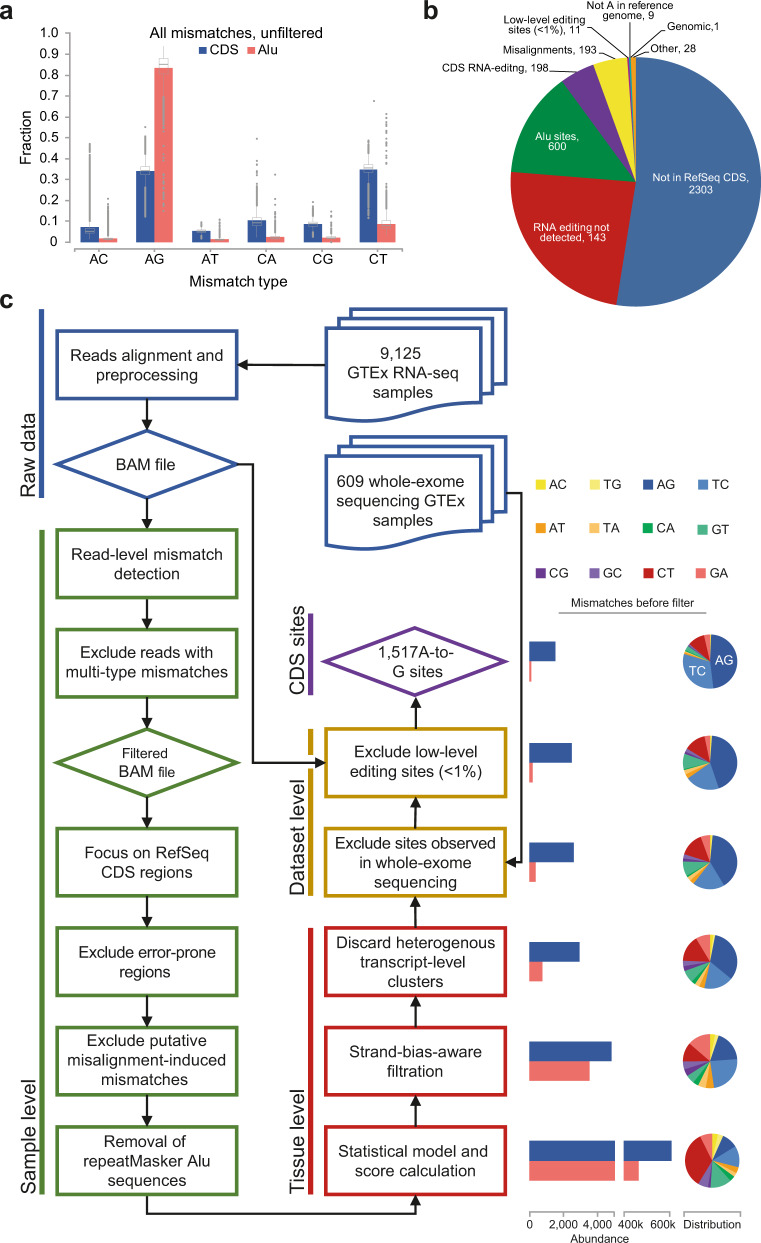
Fig. 1. A high-precision CDS-focused RNA editing detection pipeline.
a Relative abundance of all six types of RNA-DNA mismatches (strand-insensitive, i.e., A-to-C includes also T-to-G, etc.) following an alignment of 9125 GTEx RNA-seq samples to the reference human genome. All mismatch events are included, with no filtering. Multiple mismatches to the same genomic position are counted as separate events. Enrichment of A-to-G mismatches, presumably due to A-to-I RNA editing events is readily detectable within Alu elements. However, no such enrichment is observed in Alu-free CDS, where the editing signal is dwarfed by the noise. Colored bars and box-and-whisker plots represent the mean and the full distribution, respectively, of the relative abundances. See Supplementary Data 1 for the number of biologically independent samples per tissue. b Classification of all A-to-I sites annotated in REDIportal12 database as CDS. Merely 198 out of 4386 sites (4.5%) were detected by our pipeline as reliable CDS RNA editing sites while the majority of the sites were excluded from our analysis due to the reasons indicated in the panel. c A flowchart summarizing the main steps of our CDS RNA editing detection pipeline. Briefly, 9125 GTEx RNA-seq samples from various donors and tissues were aligned to the reference genome and DNA–RNA mismatches were detected and filtered within each sample separately. Results were aggregated for each tissue type for further filtration steps. Finally, the resulting candidate sites were filtered using global dataset criteria to yield a final 1517 reliable CDS A-to-I RNA editing sites (see “Methods” for details). Rightmost panel shows the mismatches abundance and distribution before each of the final filtering steps, demonstrating the increase in signal-to-noise ratio per step. Source data are provided as a Source Data file.