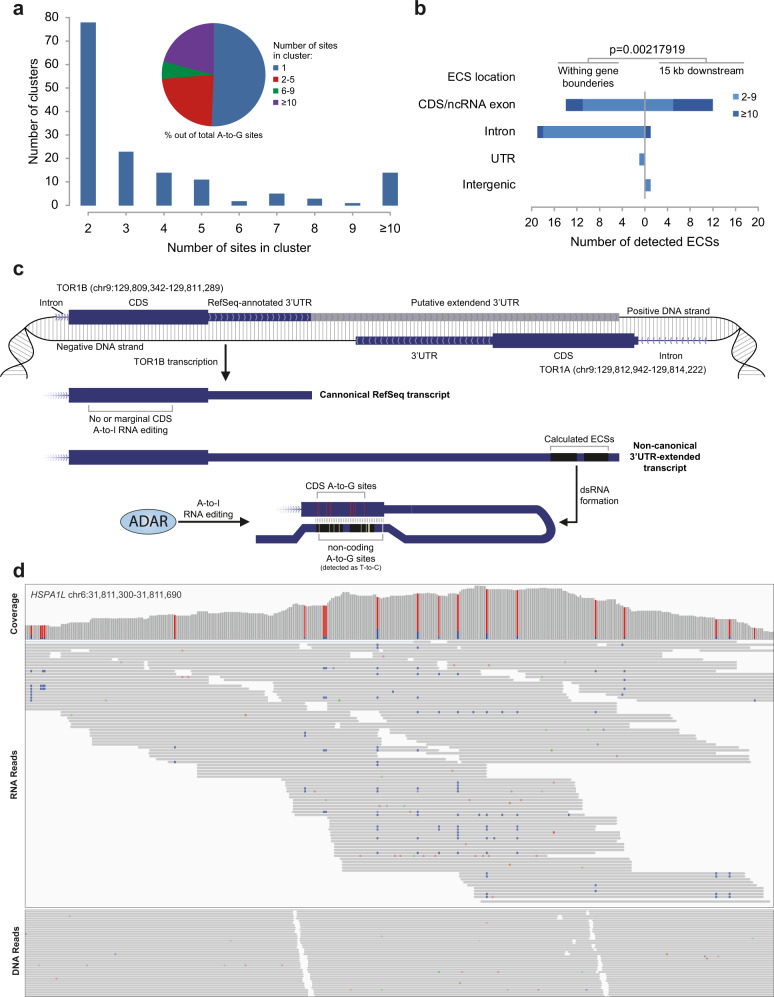
Fig. 4. Many of the CDS editing sites are clustered.
a Bar plot: Distribution of editing clusters by size. Pie chart: Relative abundance of editing sites by cluster size. b Editing requires a dsRNA secondary structure, often depending on a distant editing complementary sequence (ECS). Distribution of putative ECSs location (“Methods”) shows that smaller clusters tend to depend on an intronic ECS, while the ECS for large clusters (≥10) is often found out of the annotated gene boundaries, overlapping an exon of a neighboring downstream gene. c Editing due to a downstream reversely-oriented paralog gene, overlapping an extended 3′UTR. The TOR1B gene contains a 10-site RNA editing cluster in its coding sequence. The corresponding ECS is a closely-related sequence that overlaps a coding exon within the neighboring paralog TOR1A gene (genomic coordinates of the last exon of both genes are shown). Presumably, the 3′UTR of edited TOR1B transcripts is longer than that of the Refseq canonical transcript, extending to overlap the coding sequence of Tor1a (on the opposite strand). These extended-3′UTR variants of the Tor1b transcripts contain two highly-similar reversely-oriented sequences, and can form a long and stable dsRNA structure, resulting in extensive editing of both the CDS (red lines) and the TOR1A-overlapping 3′UTR (yellow lines). A 14-site cluster of T-to-C sites is detected in the coding sequence of TOR1A, a hallmark of A-to-I editing in antisense transcripts (see Fig. 2d). See Supplementary Data 3 for further evidence for expression and editing of the extended UTR of TOR1B. d Editing at one of the large clusters within the HSPA1L transcript. Up: pile-up of the RNA reads’ coverage, 19 different editing sites are observed in this 391 bp-long segment of the editing cluster (red and blue bars stand for A and G fractions, respectively). Bottom: Matched RNA and DNA sequences (presented is the pooled data for the three individuals, for simplicity). Editing events (A-to-G mismatches) are shown in blue (different colors represent other type of mismatches). The absence of A-to-G mismatches in the matched DNA samples and lack of linkage between neighboring sites both support the sites being RNA-edited.