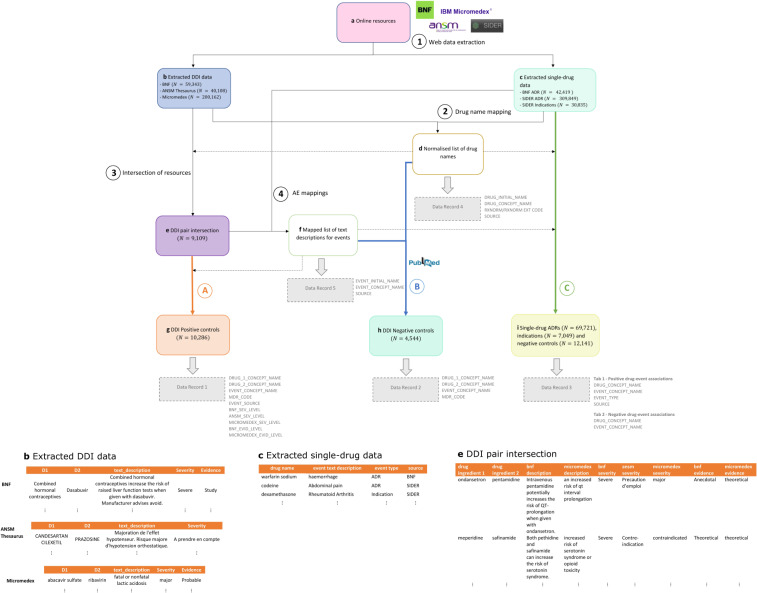
Fig. 1.
Data analysis workflow to generate positive and negative controls from DDI online resources with associated evidence for their component drugs. (1) DDI and single-drug online resources data (a) are extracted and stored to separate tables (b,c). (2) Drug names are normalized (d). (3) The intersection of the DDI online resources is extracted to a different table (e) and (4) English language text descriptions (for DDIs and single-drug ADRs) are annotated for AEs (after drug name masking) (f). (A) DDI pairs from (e) are assigned AEs based on the description mappings (g). Positive controls are published in Data Record 1. (B) Negative controls are generated using drugs and AEs from (g), ensuring that drug pairs cannot be found in (b) or in PubMed using a customized query (h). Negative controls are published in Data Record 2. (C) The filtered set of drugs from (d) is linked to AE and indication concepts using available evidence from (c) and negative controls for single drugs are generated following a similar process to the one described above for DDIs (i.e., drug-event is not an ADR mentioned in (c) or in PubMed using a customized query) (i). ADRs, indications and negative controls for single drugs are published in Data Record 3. Data Records 4 and 5 contain mappings of drug names and text descriptions for events, respectively. Column headers appear in grey font next to each Data Record box. Sample records from (b), (c) and (e) can be found in the bottom part of the figure.