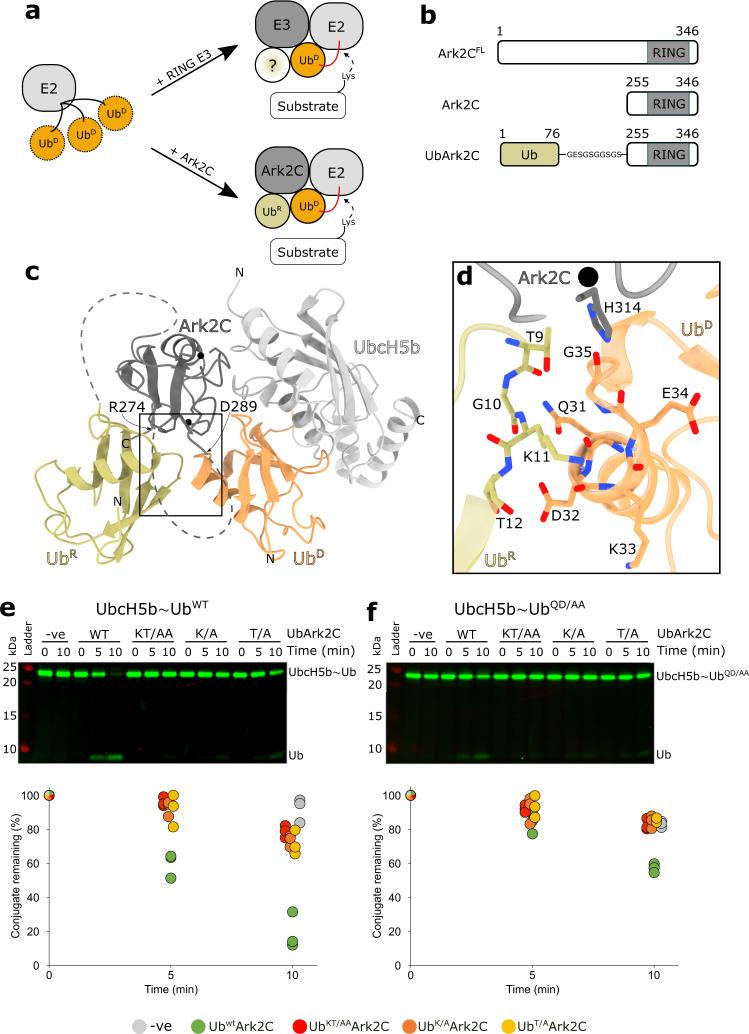
Fig. 1. Structure of the Ark2C catalytic complex and analysis of the UbR–UbD interface.
a Cartoon illustrating how RING E3 ligases stabilise the closed conformation of the E2~Ub conjugate (top) and the role of UbR for Ark-like RINGs. b Schematic showing the domain architecture of Ark2C and the constructs used in this study. c Cartoon representation of the activated complex of UbArk2C–UbcH5b~Ub; Ark2C (dark grey), UbcH5b (pale grey), UbR (yellow) and UbD (orange). Black spheres depict zinc ions. d Close-up showing the contacts between UbR and UbD as sticks. Red and blue represent oxygen or nitrogen atoms, respectively. Carbon atoms are coloured as in (c). e Discharge assays assessing the ability of the indicated UbArk2C proteins to hydrolyse the WT UbcH5b~Ub conjugate. f Discharge assays as in (e) except Gln31 and Asp32 in ubiquitin conjugated to UbcH5b were mutated to Ala, referred to as UbcH5b~UbQD/AA. In both (e) and (f) ubiquitin used to prepare the UbcH5b~Ub conjugate was labelled with Cy3 and all gels were imaged and quantified using Image Studio Lite (Li-COR Biosciences). Experiments were performed in triplicate using 0.25 μM E3 ligases and 0.125 mM l-lysine. All repeats were quantified and are shown. Source data are provided in the Source Data file.