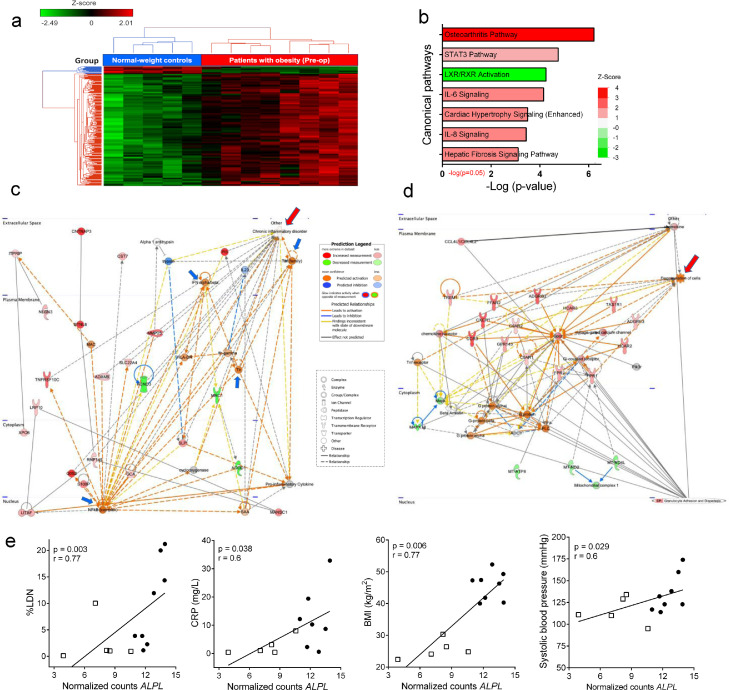
Figure 2.
Differential gene expression between MOP and NWC. (a) Dendrogram and Heat-Map for Unsupervised Hierarchical Clustering in 8 MOP and 5 NWC, based on the 238 DEG. Each column represents a MOP or NWC, and each row represents a single gene. Red and green colors denote increased and decreased gene expression, respectively. Data were z-score normalized. In the color bar above the heatmap, red indicates the 8 patients' group, and blue indicates the 5 individuals from the control group. (b) Top significantly enriched canonical pathways (FDR<0.1, FDR-unadjusted p-value<0·05). The significance of the enrichment is shown on the x-axis, and the bars are colored based on the z-score predicted activation (positive z-score, red) or pathway inhibition (negative z-score, green). (c) Top de-novo network predicted by IPA ‘Cell-To-Cell Signaling and Interaction, Dermatological Diseases and Conditions, Organismal Injury and Abnormalities’ enriched by DEG and their potential interplay (score of 37). The most significant specific disease term highlighted in this network is chronic inflammatory disorder (p=4·98E-06, associated with 19 of 35 genes), denoted with a top-right red arrow. (d) Second de-novo network predicted by IPA indicating enrichment with molecules connected to degranulation of cells, indicated with a top-right red arrow. Nodes represent genes and lines show the relationship between genes. Nodes in red are upregulated in MOP versus NWC, and nodes in green are downregulated. The color intensity represents the relative magnitude of change in gene expression. Orange or blue nodes are predicted to be activated or inhibited genes, respectively, based on the downstream state of other molecules in the network. White nodes have a z-score of 0, indicating neither activation nor inhibition, and those in gray have no predicted activity pattern. Lines are solid or dashed to indicate direct and indirect relationships, respectively. Orange lines indicate a relationship leading to activation, blue lines leads to inhibition, and yellow lines indicate that the downstream state of the genes is inconsistent with the expected relationship. Lines in gray show the interaction but have no predicted effect. Blue arrows are indicating TNF, IFN-alfa/beta, NF-kB and TLR nodes. (e) Correlation between RNA normalized counts of ALPL with %LDN, CRP, BMI, and systolic blood pressure. MOP are represented with black circles and NWC with white squares. The results are presented as Spearman 's correlation coefficient (r) and p-value. Results were considered significant with a p-value ≤ 0.05. MOP indicates morbidly obese patients; NWC. Normal-weight control; DEG, differentially expressed genes; IPA, ingenuity pathway analysis; TNF, tumor necrosis factor; IFN; interferon; NF-kB, nuclear factor kappa B; TLR, toll-like receptor; ALPL, alkaline phosphatase; LDN, low-density neutrophils; CRP, C-reactive protein; BMI, body mass index; FDR, false discovery rates.