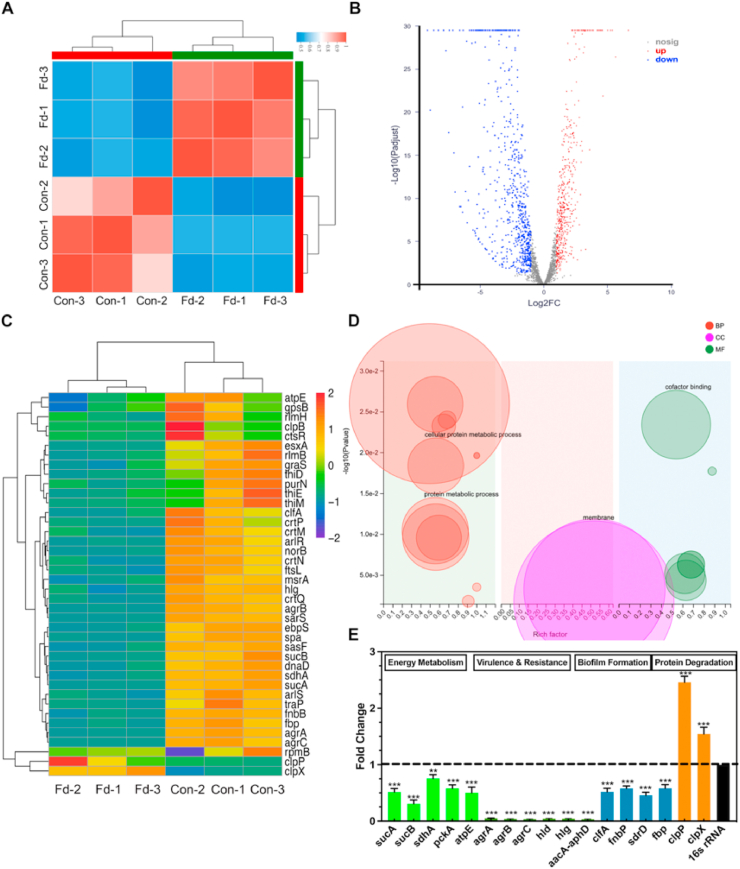
Fig. 3.
Changes in the transcriptome of MRSA treated with felodipine or DMSO (control). (A) Heatmap of correlation between samples. The different colors of the squares represent the different correlation coefficients of the two samples. (B) Volcano map of differentially expressed genes (DEGs). Each dot represents a specific gene. Red indicates significant upregulation, blue indicates significant downregulation, and gray indicates non-significant differential expression. (C) Clustering heatmap of some important DEGs. The shade of color indicates the amount of gene expression. Each group contains data for three independent samples. (D) Go enrichment analysis of DEGs. Each bubble represents a GO Term. The size of the bubble is proportional to the number of genes enriched to the GO Term. The different colors represent the three major classifications of GO. BP = biological process; CC = cellular component; MF = molecular function. (E) Validation of gene expression by RT-PCR. Felodipine inhibited the gene expression associated with energy metabolism, biofilm formation, aminoglycosides resistance, and bacterial virulence. And felodipine increased the gene expression associated protein degradation. Data analysis was performed using the comparative CT method, with 16S rRNA serving as the comparator. The results are presented as fold-changes relative to the control, which was set to a value of 1. Data are expressed as the mean ± SD. n = 3; **p < 0.01; ***p < 0.001.