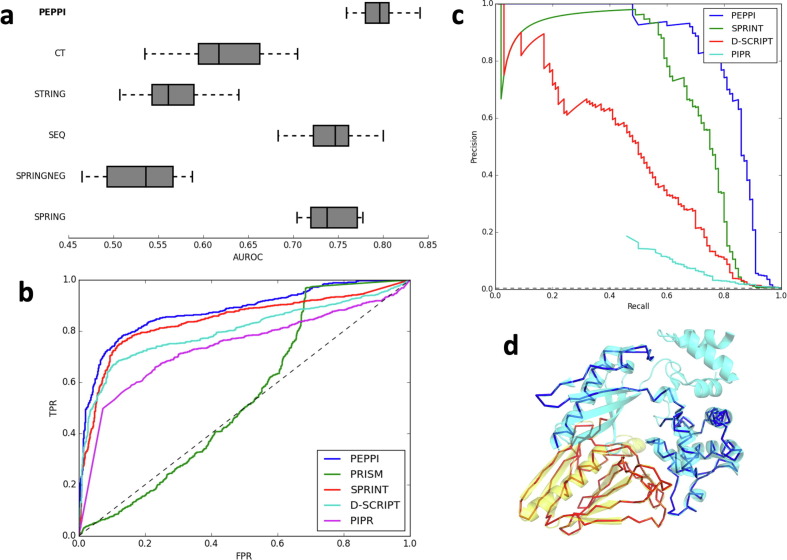
Figure 2.
PEPPI benchmark results. (a) 10-fold cross validation AUROC reveals that the full PEPPI pipeline outperforms its component modules: the neural network classifier (CT), the functional association data (STRING), the sequence similarity method (SEQ), the structure similarity method (SPRING), and the non-interaction similarity method (SPRINGNEG). (b) An ROC curve of the performance of PEPPI against PRISM, a structure similarity-based method, SPRINT, a sequence similarity-based method, PIPR, a deep learning-based method, and D-SCRIPT, a structure-aware deep learning-based method on a balanced testing set. The dotted line represents the performance of random classification. (c) Precision-recall curve of the performance of PEPPI against several other comparable programs on an unbalanced testing set. The dotted line represents the performance of random classification. (d) A superposition of an example dimer model (PDB 3CI0; chain J in red, chain K in blue) on its dimer template structure (PDB 5VTM; chain W in yellow, chain X in cyan).