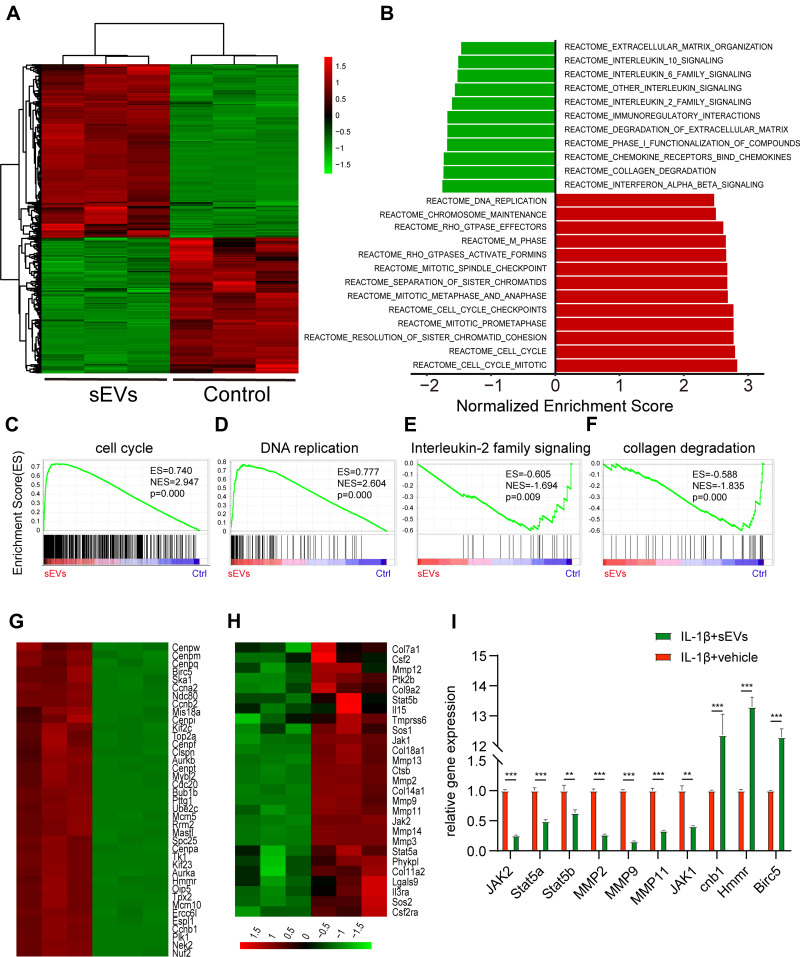
Figure 5.
iMSC-sEVs Modulate the Gene Expression Pattern of Rat Tenocytes Under the Stimulation of IL-1β. (A) Heatmap representing the hierarchical clustering of significant differential expression genes in rat tenocytes treated with or without iMSC-sEVs. (B) GSEA analysis showing canonical pathways of the differentially expressed genes in tenocytes. (C–F) GSEA analysis identifies enrichment score for modules of cell cycle (C), DNA replication (D), interleukin-2 family signaling (E), and collagen degradation (F) in tenocytes after iMSC-sEVs treatment. Heatmap representation of differentially expressed genes (fold change >2) in the GSEA analysis associated with cell cycle (G), interleukin-2 family signaling and collagen degradation (H). (I) RT-qPCR analysis of anti-inflammatory gene expression in tenocytes after iMSC-sEVs treatment. The experiment was repeated three times independently. P-value is indicated as sEVs group versus ctrl group. **P < 0.01, ***P < 0.001.