Figure 6. Circadian deregulation in human HCCs.
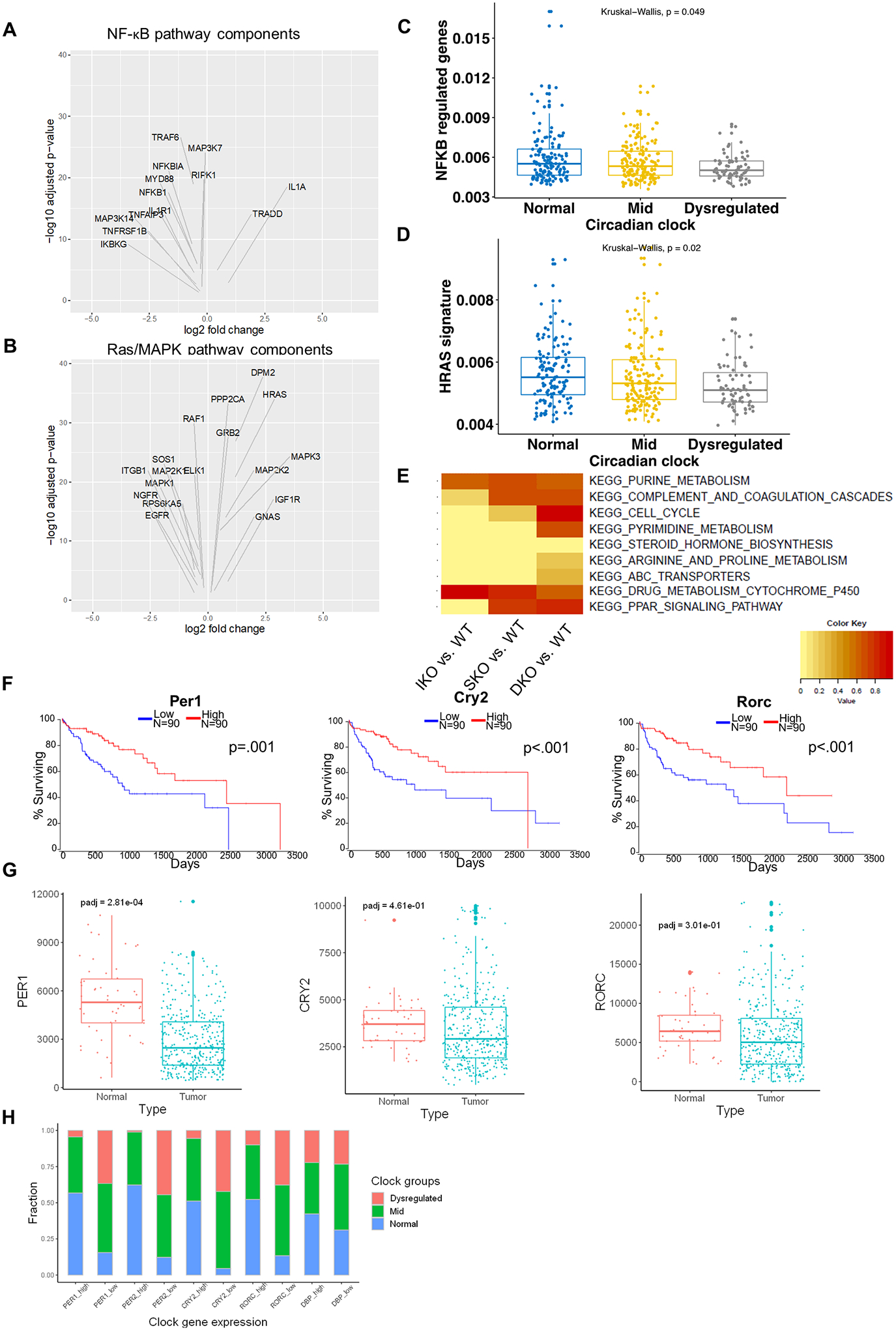
(A) Volcano plots of differentially expressed NF-κB pathway components in clock-dysregulated human HCCs (adjusted p<0.05). (B) Volcano plots of differentially expressed Ras/MAPK pathway components in clock-dysregulated human HCCs (adjusted p<0.05). (C) NF-κB target gene expression profiles in human HCCs with Normal, Mid (<5 clock genes disrupted), or Dysregulated (>5 clock genes disrupted) circadian status. (D) HRas target gene expression profiles in human HCCs as stratified in C. (E) Enrichment for KEGG pathways previously identified to correlate with disrupted circadian processes in human HCC. DEGs from hepatocytes isolated from 2-month-old livers were analyzed to determine similarity to this “circadian-disrupted” signature. (F) Association between clock gene expression and overall survival in HCC patients. TCGA-LIHC cohort data were analyzed and Kaplan-Meier plots were generated using OncoLnc; upper and lower quartiles of gene expression were used to determine “high” and “low” expression, respectively. (G) Normalized reads of the indicated circadian transcripts in normal (non-tumor liver) and tumor (HCC) tissues in patients from the TCGA-LIHC cohort. Box-and-whisker plots indicate quartiles of expression for each gene. (H) Fractions of patients with Normal, Mid, or Dysregulated circadian status in patients with upper-quartile (High) or lower-quartile (Low) expression of the indicated clock gene.
