Figure 2. MALT1 expression is linked to EMT in GPCR+ breast cancer.
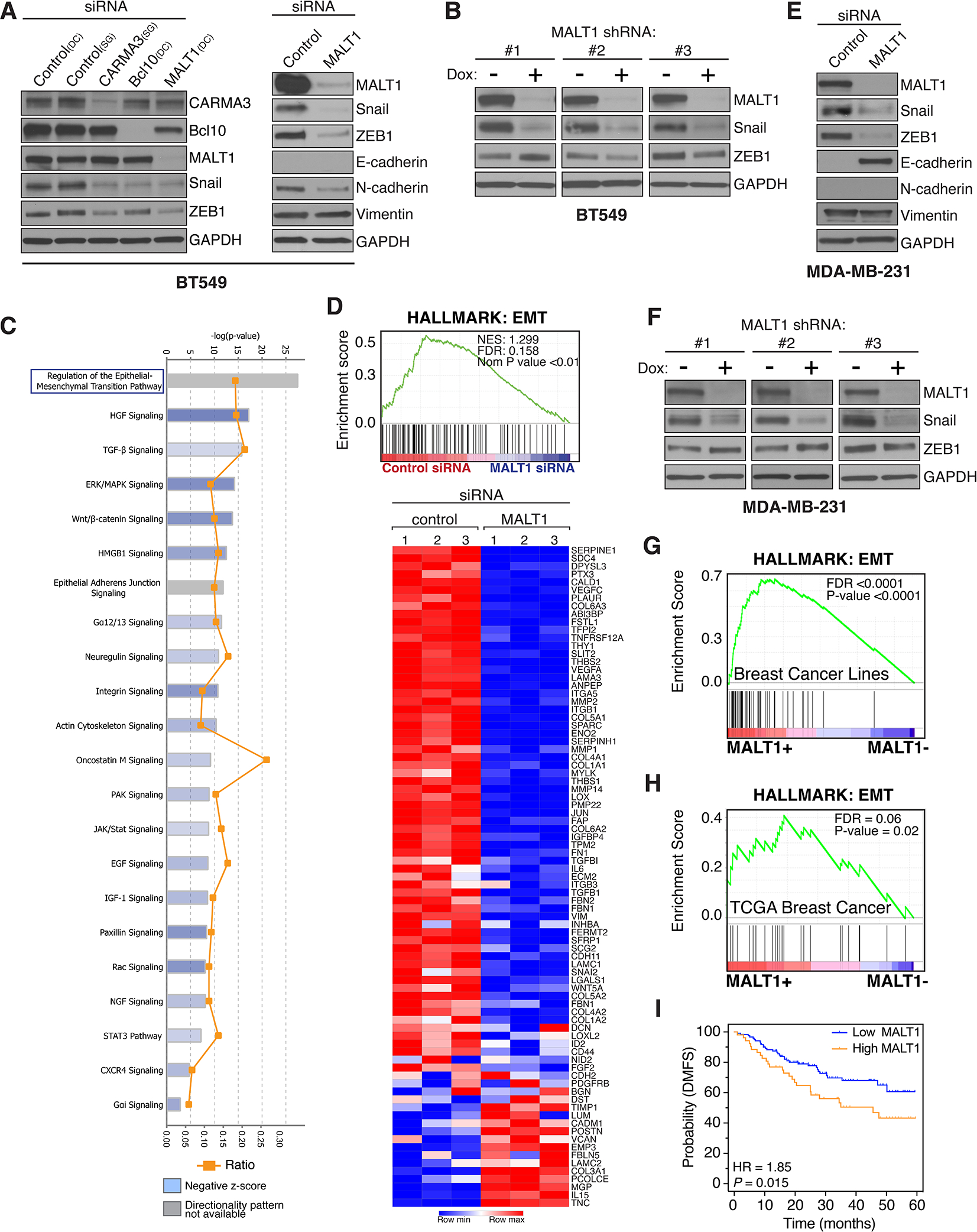
A and B, Effect of CARMA3, Bcl10, or MALT1 knockdown, in AT1R+ BT549 cells, on markers of EMT. Knockdown was accomplished by either transient siRNA transfection (A) or by stably-integrated doxycycline-inducible shRNAs (B). In the case of shRNA-mediated knockdown, cells were treated for 5 days with 2 μg/ml doxycycline to induce shRNA expression prior to harvesting and immunoblot analysis. DC, Dharmacon siRNA; SG, Sigma siRNA. C and D, IPA and GSEA linking MALT1 levels with EMT in BT549 cells. Cells were subjected to MALT1 knockdown using transient siRNA transfection, in biologic triplicate, and gene expression profiles were subsequently analyzed using the NanoString Pan-Cancer Progression Panel. Gene expression data was analyzed by IPA to identify pathways impacted by the loss of MALT1, revealing EMT as the top-most significantly affected pathway (C). The same data was evaluated using GSEA and the Hallmark EMT signature (D). The heatmap shows individual genes included in the Hallmark EMT gene set and their specific alterations in response to MALT1 knockdown. E and F, Effect of transient (E) or stable (F) MALT1 knockdown, in PAR1+ MDA-MB-231 cells, on markers of EMT. Analyses were carried out using an approach identical to that used for BT549 cells (panels A and B). G, GSEA demonstrates an association between MALT1 expression and the EMT Hallmark signature in breast cancer cell lines included in the Hoeflich dataset (37). H, GSEA demonstrating an association between MALT1 expression and the EMT Hallmark signature in TCGA breast cancer cases. I, High MALT1 expression is associated with worse DMFS in Grade-3 TNBC with 5 years of follow-up. Dataset from Gyorffy et al (73) and analyzed using KM-plotter (www.kmplot.com).
