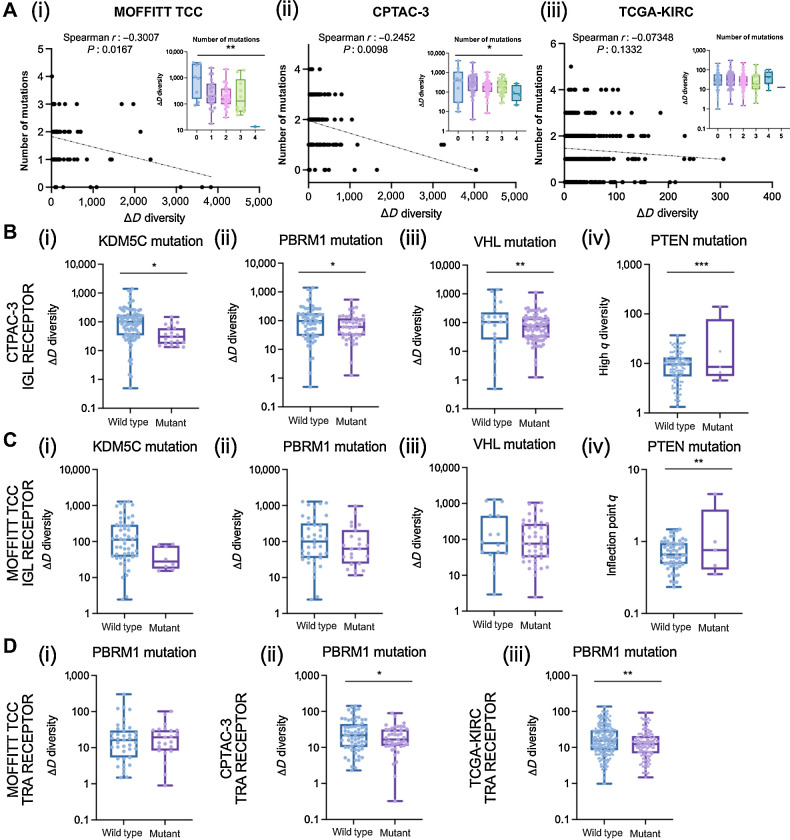
Figure 5.
Associations of diversity metrics with mutational landscape in ccRCC. A, Number of mutations were negatively correlated with CDR3 recovery richness. Spearman correlation coefficients between number of mutations and ΔD diversity were calculated for the (i) Moffitt TCC total (TRs+IGs) recoveries (Spearman r: −0.3007, P: 0.0167; ANOVA, P: 0.0025); CPTAC-3 total recoveries (Spearman r: −0.2452, P: 0.0098; ANOVA, P: 0.0213), and (iii) TCGA-KIRC total available (TRA+TRB) recoveries (Spearman r: −0.07348, P: 0.1332; ANOVA, P: 0.3905). B, IGL recoveries had reduced richness in CPTAC-3 patients with (i) KDM5C mutations (mean score of wild type was 167.9 and mutant was 44.41; P: 0.0298), (ii) PBRM1 mutations (mean score of wild type was 182.2 and mutant was 92.06; P: 0.0421), and (iii) VHL mutations (mean score of wild type was 247.7 and mutant was 115.0; P: 0.0094) and increased evenness in patients with (iv) PTEN mutations (mean score of wild type was 10.61 and mutant was 35.35; P: 0.0001). C, IGL recoveries had reduced richness in Moffitt TCC patients with (i) KDM5C mutations (mean score of wild type was 248.1 and mutant was 42.96; P: 0.0906), (ii) PBRM1 mutations (mean score of wild type was 259.8 and mutant was 156.4; P: 0.2200), and (iii) VHL mutations (mean score of wild type was 307.7 and mutant was 190.4; P: 0.1993) and increased evenness in patients with (iv) PTEN mutations (mean score of wild type was 0.7323 and mutant was 1.427; P: 0.0083). D, TRA receptor richness was (i) not different in Moffitt TCC patients with PBRM1 mutations (mean score of wild type was 32.03 and mutant was 24.59; P: 0.5397), (ii) decreased in CPTAC-3 patients with PBRM1 mutations (mean score of wild type was 33.63 and mutant was 22.53; P: 0.0470), and (iii) decreased in TCGA-KIRC patients with PBRM1 mutations (mean score of wild type was 23.84 and mutant was 17.06; P: 0.0032). Unpaired t tests were used to compare two group data and ANOVA was used to compare grade, three group data. *, P < 0.05; **, P < 0.01; ***, P < 0.001.