Figure 3. Chk1 inhibitors disrupt STAT3 activation in MM cells.
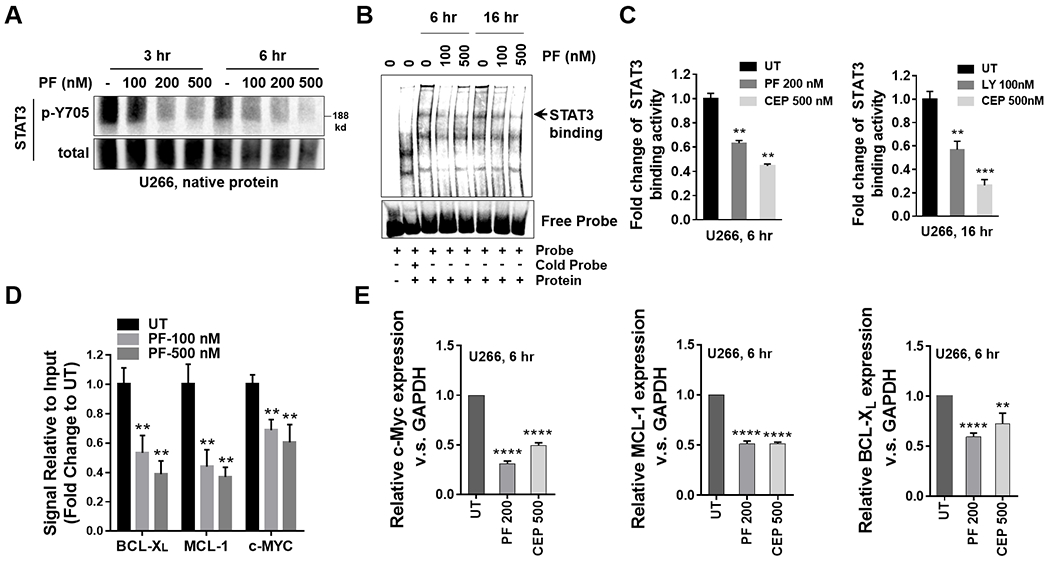
A, U266 cells were treated with indicated concentrations of PF for 3 and 6 hr. Native nuclear protein extracts were prepared and were separated on a 6% native PAGE gel. Electrophoresis was performed in the absence of SDS. Proteins were immunoblotted with p-Y705 STAT3 and total STAT3. Total STAT3 served as a loading control. B, Nuclear extracts from U266 cells treated with the indicated concentrations of PF for 6 and 16 hr were subjected to EMSA analysis. C, U266 cells were treated with indicated concentrations of PF, CEP or LY for 6 and 16 hr. A STAT3 DNA-binding ELISA assay was employed to evaluate the ability of STAT3 in cellular nuclear extracts to bind to its corresponding consensus sequence that had been immobilized on a 96-well plate. D, U266 cells were treated with the indicated concentrations of PF for 16 hr. ChIP experiments were performed to detect the presence of STAT3 at the promotors of the BCL-XL, MCL-1, and c-Myc genes after treatment with PF. Data was normalized with non-immune IgG (negative control) and STAT3 antibody, and the relative fold- change (vs untreated controls; UT) is presented. E, Relative mRNA expressions of c-Myc, MCL-1 and BCL-XL were determined by real-time reverse transcription-PCR analysis. GAPDH served as an internal control. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
